Removing Dark Region Artifacts using Deep Neural Networks with a Hyperparameter Search
ABSTRACT
This study aims to use machine learning in the form of Deep Neural Networks (DNN) to remove unwanted off-axis signals that causes dark region artifacts (DRAs) to appear on ultrasound images of dense, bright bodies, such as kidney stones. DRAs cause dark areas to appear around the bodies in ultrasound images formed through beamforming. This was done by creating 27 different DNNs with varied parameterization of drop out, hidden layers, and batch size in order to see whether the DNNs were effective at removing the DRAs and if so, what parameters were the most effective. After data analysis using MATLAB Excel to calculate and analyze both image contrast and contrast to noise ratio, it was concluded that the all 27 DNNs were successful at removing at least some of the DRAs while having low dropout was the most conducive to creating the best image. This is important as ultrasounds are a inexpensive non-invasive way to examine tissue, and more complete and accurate images are vital for proper diagnosis and understanding of that tissue.
INTRODUCTION.
Ultrasound uses reflection of sound waves to produce images. This is commonly used in imaging tissue as a non-invasive way to examine growths, such as tumors or kidney stones. The sound signals are made into images through a process known as beamforming, in which an ultrasound probe with several different sensors combines their individual information to form one coherent image through an equation that waits for the data from each sensor and adds it up (delay and sum). However, the sound does not always reach the channels after being reflected off the tissue flawlessly. The sound often returns with extra data known as clutter, commonly caused by off-axis signals, distortion of the sound, or reverberation, the sound echoing off the tissue. This clutter makes the image blurry after assembling, known as image-degradation. To combat the degradation and improve image quality, adaptive beamforming, which changes the equation depending on the characteristics of the image, is beginning to replace traditional delay and sum beamforming [1].
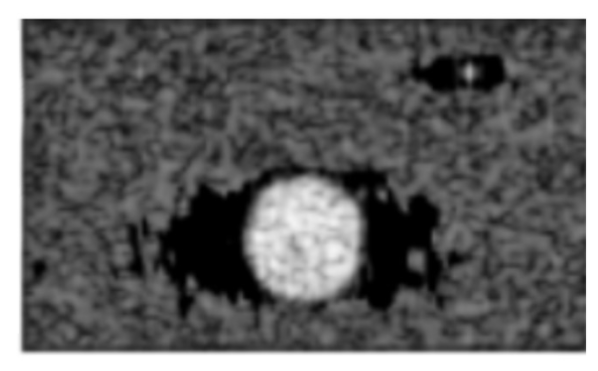
Currently, one major result of using an adaptive beamformer is strong off-axis signals in the regions surrounding a region of interest, resulting in lowered coherence, or lack of information [2]. Dark region artifacts (DRAs) are a very common phenomenon resulted from off-axis signals in adaptive beamformers, and which limit adaptive beamformers from being used in real world settings [3]. DRAs are a phenomenon created by some adaptive beamformers wherein dense objects, such as kidney stones, reflect sound much better than the surrounding soft tissue. As a result, measures used to weight the image are degraded which results in dark spots, which typically suggest an absence of tissue as shown in Figure 1. This makes it extremely difficult for healthcare providers to fully assess the condition of the surrounding tissue.
Figure 1. Example of a cyst with dark region artifacts surrounding the cyst on the horizontal. (Units in mm).
Recently, initial research has demonstrated the benefit of using deep neural networks (DNNs) to reconstruct ultrasound images, but the DNN training is still a big challenge [1]. DNNs work by learning what data is good data and learning what is bad data. It compares its results for filtering to idealized simulated removals to learn what it can do better. Previous studies have shown that using linear ultrasound simulation tools to generate training data for reconstructing ultrasound images is helpful, but limited image quality is one concern of this approach. Surprisingly, previous studies have shown that assignment of different settings for the adaptive beamformers known as hyperparameters for DNN beamformers significantly improved image quality [1]. Furthermore, pilot experiments suggested that 3 parameters were found to be crucial for the quality of images created by the DNNs: hidden layers (the number of steps that the DNN takes), dropout (what percentage of the data is being dropped to prevent overfitting) and batch size (how many examples the DNN is being trained with). The current study aims to broaden the use of DNNs to reduce DRAs by first removing off-axis data before using an adaptive beamformer to avoid the signals from being used to create the image. Additionally, this study attempts to determine which parameter was the most critical in removing off-axis signals while still preserving the speckling, or pattern around the cyst.
MATERIALS AND METHODS.
27 DNNs were designed in Python using Anaconda and PyTorch packages. Each DNN had different parameters. Three parameters were adjusted in this study: hidden layers (1, 4, or 8); dropout (0, 0.25, or 0.5) and batch size (32, 128, or 512). A fully connected neural network architecture with a rectified linear unit (ReLU activation function), a way for the DNN to know if it is successful) will be used. The learning rate was adaptively tuned using the adaptive moment estimation method, with a learning rate (α) of 10-3, and averaging coefficients of β1=0.9 and β2=0.999.
The DNN was then trained using simulated data. Ten simulated ultrasonic phantoms were generated using Field II simulation software in Matlab. Eight were used for training and another two were used to assess error after training. These phantoms mimic a region of tissue with a bright cyst with a darker background. The cysts were scaled to be 30 dB larger in amplitude than the tissue, allowing the cysts to be a lot brighter. Additional noise was added at -10 dB relative to the tissue to simulate reverberation that would occur naturally.
Simulated data with the cysts were modelled after a linear L7-4 that was acting as a transducer, or the ultrasound probe that would usually be put against the body. The data was simulated in an environment similar to a human body. The probe was designed to have 65 individual sensors, or elements with a size of 0.25 mm. The speed of sound was assumed to be 1540 m/s, which is the approximate speed of sound as it travels through soft tissue.
Training is performed by exposing the DNN to curated pairs of channel data (input) and manually-corrected channel data (output). The manually-corrected channel data was formed by excluding the strong off-axis signal signals produced by the cyst that are normally superimposed on the background. This correction can only be performed in simulation, where the cyst and background data are generated separately. In clinical imaging, these contributions are not easily separable, which motivates using simulated data for the development of a DNN correction methodology.
During training, the DNN develops an algorithm that accurately recreates an image of the cyst without DRA from the original data. The final algorithm is optimized through a series of epochs, cycles where a DNN makes a slight adjustment to the algorithm. A successful change is determined through mean square error by comparing the resulting image with the actual image of the cyst without DRA. The training was terminated if the mean square error did not improve after 20 consecutive epochs, and the last successful epoch is used. This training model has been previously used by Luchies and Byram. [1]
Whether the DNN successfully removed, the DRA was determined by a measurement of contrast. Final images were formed from the post-processed channel data using the coherence factor adaptive beamformer, which suffers from DRAs. Coherence factor images are formed by multiplying the delay-and-summed image by a weight determined by the signal coherence. This is performed as
\[CF\ Image=\frac{\left|\sum_{i=0}^{N-1}S\left(i\right)\right|^2}{N\ \sum_{i=0}^{N-1}\left|S\left(i\right)\right|^2}\sum_{i=0}^{N-1}S\left(i\right)\tag{1}\]
where S(i) is the signal on channel i after focusing delays are applied and N is the total number of channels available [4].
Contrast was calculated by comparing the area that previously contained a DRA with an area of the background without a DRA, using the following formula
\[Contrast=10\ast log10\left(\ \ \frac{\mu_{body}}{\mu_{background}}\right)\tag{2}\]
where μ indicates the mean image intensity. If the contrast is very low and close to 0, the DNN is defined as successful removal of the off-axis signals, suggesting a uniform tissue pattern is achieved. Another metric, contrast to noise ratio (CNR) was used as a backup for contrast, as sometimes contrast can end up being infinity if the background is completely black [5]. CNR is calculated according to the following formula
\[CNR=10\ast log10\ \left(\ \frac{\mu_{body}-\mu_{background}\ }{\sigma_{background}\ }\right)\tag{3}\]
where σ indicates the standard deviation.
RESULTS.
DNNs effectively removed contrast between DRA and other background.
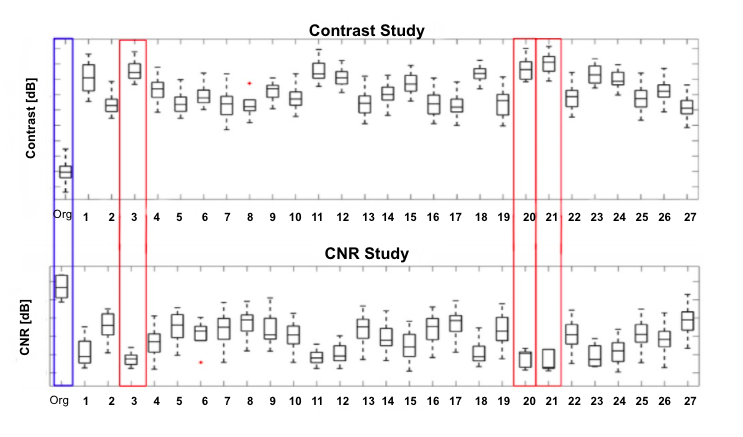
Removal of these dark regions in ultrasound images will provide more detailed information for clinical doctors. To determine whether utilizing DNNs will improve the image contrast, the algorithms obtained through training were tested with 5 new images with DRAs. Unlike in training, it was not exposed to what the images were supposed to look like without the DRAs. Then, the contrast and contrast to noise ratio of both the original images and the images after being processed by the DNN were made into a box and whiskers plot (Fig. 2), which shows how the DNNs had metrics closers to 0 than the original, suggesting improvement. The contrast measures were then log compressed and averaged between each DNN to be analyzed. Compared to the average contrast of the original images (which is 0.977), the average contrast of all 27 different DNN conditions was significantly increased. In contrast, the Contrast-Noise Ratio (CNR) of all 27 different DNN conditions were significantly decreased as shown in figure 2. These data suggest that using DNNs to reconstruct a ultrasound image may improves image contrast. Of note, the three top CNNs conditions are 21st, 20th, and 3rd when analyzed for contrast.
Figure 2. Contrast and CNR of each of the DNNs, with the best DNNs are boxed in red.
DNNs effectively limited dark region artifacts.
One drawback of most adaptive beamformers is dark region artifacts (DRA), which may mislead clinical doctors. Often, these metrics do not tell the whole story and the images still have to be hand selected. It is important that the human brain can see the same improvement that the numbers do. Figure 3 shows the difference between the original cyst, the cyst after being processed by the best DNN and the same cyst after being processed by the worst DNN. When compared with the original image, which had prominent dark region artifacts, even the worst DNN improved the imaging of the area around it. However, there was still a visible darker patch in the DRA. This was changed in the best DNN. It preserved the speckling the best while being able to completely fill out the dark region artifacts.
Figure 3. The original cyst and compared to the same cyst after being processed by the best and worst DNNs. Both DNNs show removal of the black area that surrounds the cysts in the original image.
Parameter Effects.
The 27 DNNs were created with different combinations of three parameters (Hidden layers, Dropout and Batch size), and contrast of the images was quantified by Matlab.
The parameters of each of the DNNs were lined up with the ranking of the DNNs by contrast, and a correlation coefficient was found between each of the 3 alters parameters (hidden layers, dropout, batch size) and the contrast. There was a weak correlation between both the batch size and the hidden layers, with correlation coefficients of -0.200 and -0.343 respectively. However, the dropout had a higher correlation coefficient at 0.569, showing a possible correlation between lower dropout and improved artifact removal. This possibility is supported by how 6/7 of the best DNNs in respect to contrast had the same dropout of 0. Table 1 shows the parameters for each of the DNNs and the subsequent contrast.
Table 1. Average contrast of 27 DNNs, measured utilizing Matlab after adjustment of three parameters. Lower contrast indicates the most improvement.
DISCUSSION.
Overall, when tested on new data that it had not been previously exposed to, the DNNs were very effective in reducing the DRAs that resulted from the off-axis signals resulting from the beamforming techniques. Passing the channel data first through the DNNs before beamforming resulted in a similar contrast and CNR between the area that previously had the DRA and an unaffected part of the background. This process improved coherence and created better and more accurate images. This study also showed that the dropout could be the most important parameter for removing DRA, as low dropout had the strongest correlation with the contrast as well in addition to 6/7 of the best DNNs having no dropout. This should be tested in the future to show whether the dropout itself is what creates a DNN that is the best at removing DRA as well as testing low dropout, a measure meant to prevent overfitting, with large training sets to see if that affects the results as well. Finally, now that it is clear that a DNN can successfully remove DRA from the images, it is important to see how well this DNN works with DNNs that serve other purposes for ultrasound beamforming, as the ultimate goal is to have a DNN that can work real time to solve a multitude of issues common in ultrasound imaging. This would allow the DNN to be applied in real time would help doctors and healthcare workers around the world gain better understanding the tissue and surrounding area of dense bodies, such as kidney stones. This is especially useful because ultrasounds are one of the least expensive, non-invasive imaging techniques, as it does not require dyes or special facilities and ultrasound machines are relatively portable.
ACKNOWLEDGMENTS.
I would like to thank the School of Science and Math at Vanderbilt for the help that they have offered me in my science career and the opportunities they gave to me. I would especially like to thank Dr. Haag in being my mentor during these trying times. I would also like to thank Kathryn Ozgun for her everlasting patience and helpfulness through my entire project. Next, I would like to thank Jaime Tierney for her help with the Deep Neural Networks. Finally, I would like to thank Dr. Brett Byram and his lab for providing us with the preliminary papers and lab access for when we needed it.
REFERENCES
- Luchies, B. Byram, Training Improvements for Ultrasound Beamforming with Deep Neural Networks. Physics in Medicine and Biology. 64. (2019)
- Lediju et al, Short-Lag Spatial Coherence of Backscattered Echoes: Imaging Characteristics. IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control, 58, 1377-1388. (2011)
- Ole Marius Hoel Rindal, et al, The dark region artifact in adaptive ultrasound beamforming. 2017 IEEE International Ultrasonics Symposium, 1-1(2017).
- Li, M. Li, Adaptive imaging using the generalized coherence factor, IEEE transactions on ultrasonics, ferroelectrics, and frequency control, 50, 128-141. (2017)
- A. Rodriguez-Molares, et al, The Generalized Contrast-to-Noise Ratio. 2018 IEEE International Ultrasonics Symposium. 1-1(2018)
Posted by John Lee on Wednesday, May 19, 2021 in May 2021.
Tags: Deep Neural Network, Machine learning, Ultrasound