Optimization of 6-Thioguanosine Concentration to Detect RNA Bound by IFIT-1 via PAR-dCLIP
ABSTRACT
Interferon Induced Protein with Tetratricopeptide Repeats 1 (IFIT1) is an RNA-binding protein (RBP). IFIT1 binds to the 5’ cap of RNA and recognizes incomplete cap structures. Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Decapping and Immunoprecipitation (PAR-dCLIP) is a method in which photoactivatable ribonucleoside analogs are incorporated into cellular RNA, allowing analysis of RBP-to-RNA interactions. 4-thiouridine (4SU) is the typical analog used in PAR-dCLIP. However, uridine is not enriched in the 5’ untranslated region (UTR), where IFIT1 is known to bind the 5’ cap. 6-thioguanosine (6SG) is an analog alternative to 4SU enriched in the 5’ UTR. 6SG has cytotoxic properties, so an optimal treatment concentration must be found. A cell viability assay and High-Performance Liquid Chromatography (HPLC) were performed to determine cell viability and 6SG incorporation rate into RNA at various concentrations of 6SG. We predict greater concentrations of 6SG to yield greater cell death and incorporation.
INTRODUCTION.
RNA-binding proteins (RBPs) regulate genes at the level of RNA through post transcriptional gene regulation (PTGR) [1]. PTGR is critical to the immune system because of its role in balancing cells between a normal and immune activated state. When a pathogen is detected, the immune state is initiated, including the expression of interferon stimulated genes (ISG). Interferon Induced Protein with Tetratricopeptide Repeats 1 (IFIT1) is an ISG-RBP that recognizes the 5’ cap of RNA and distinguishes between completed cap and incomplete cap structures [2]. A cap 0 structure lacks a methyl group at the 2’ O of the first transcribed base and is considered an “incomplete cap”. A cap 1 structure possesses this methyl group and is thus considered a “complete” cap structure. Cap 0 structures exist on some viral RNAs that are bound by IFIT. This restriction of viral replication underscores IFIT1 as a classically defined antiviral protein [1].
IFIT1 binds to cap 0 structures, while the methyl group on the 2’ O of the first transcribed base of cap 1 structures prevents IFIT1 binding (Fig 1). IFIT1 binds cap 0 transcripts and sequesters the RNA from the machinery that would translate the RNA into a protein. Cap 1 structures, however, evade IFIT1 recognition and can be translated.
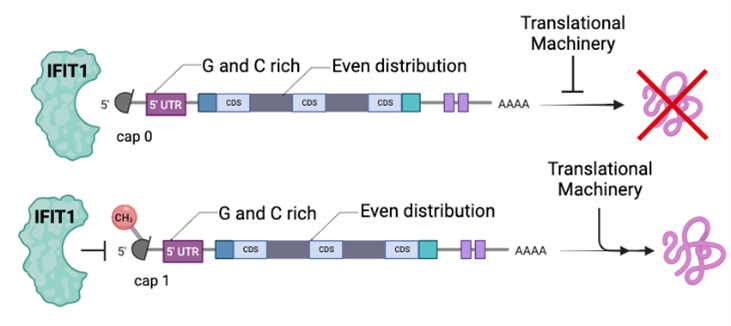
Our lab has data showing that IFIT1 is an RBP that binds cellular RNA in addition to viral RNA (Lisy et al., in preparation). Thus, we are interested in characterizing the IFIT-bound cellular transcripts to form a better understanding of IFIT1’s role in the immune system, not only as a protein involved in cellular response to infectious pathogens, but also a potential antiviral therapeutic target. Photoactivatable-Ribonucleoside-Enhanced Decapping Crosslinking and Immunoprecipitation (PAR-dCLIP) is an experimental procedure that captures RNA-protein interactions using photoactivatable ribonucleoside analogs [3]. These analogs differ from their ribonucleoside partners in that they have a sulfur group that crosslinks to aromatic protein residues when irradiated with long wave UV light [3]. The procedure starts by culturing cells in media that contains the ribonucleoside analog. The analog is then incorporated into the cellular RNA and the cells are irradiated with 365nm light. Once crosslinked to a protein it is essentially tagged. RNA can be isolated then manipulated for downstream analysis via an immunoprecipitation of the protein of interest.
The photoreactive ribonucleoside analog typically used to study IFIT1 in PAR-dCLIP is a 4-thiouridine (4SU) analog of uridine. However, IFIT1 binds to the 5’ cap along with the sections of the 5’ untranslated region (5’ UTR), which is a guanosine and cytosine rich region. This means that uridine is not abundant in this area, so using an analog not enriched in the region of interest may not reveal all possible IFIT1 to 5’ UTR binding interactions through PAR-dCLIP.
To solve this problem, this study looks at 6-thioguanosine (6SG), an analog of guanosine which is an alternative to 4SU that would be more enriched in the 5’ UTR. Using 6SG in a PAR-dCLIP with IFIT1 could provide a deeper look at IFIT1’s binding interactions. Before this analog can be used however, an updated PAR-dCLIP methods with 6SG as the analog must be established. 6SG has cytotoxic properties, so a concentration safe for cell viability must be determined. Ideally, the highest concentration that allows for viable cells and greatest incorporation into RNA is found. High Performance Liquid Chromatography (HPLC) was used for its ability to determine the number of particles in a substance, allowing varying concentrations of 6SG to be studied for their incorporation rate. We predicted that greater concentrations of 6SG will lead to increased cell death and yield greater incorporation rates compared to lower concentrations of 6SG.
MATERIALS AND METHODS.
Cell Culture and Treatment.
Human THP1 monocytes (ATCC) with the rtTA transgene were grown according to published methods [4]. Briefly, cells were cultured in RPMI (Gibco) with 105 μg/mL fetal bovine serum (FBS from Peal Serum), 100 μg/ml streptomycin (Gibco), 100 μg/ml penicillin (Gibco), 100 μg/ml hygromycin (Invivogen), in addition to 5% sodium pyruvate (Gibco).
Prior to each collection for Cell Viability or HPLC, THP1 cells were grown to a concentration of 1×106 cells/mL in 20 mL of complete THP1-rtTA media and incubated for 16hrs with 6SG (BioSynth) resuspended in 1 M NaOH (BioSynth) at increasing concentrations (1 μM, 10 μM, 100/132.1μM). A positive control of 50 μM Tamoxifen (Sigma) and negative control of no treatment were also incubated.
Cell Viability Assay.
To observe the cytotoxicity of 6SG at increasing concentrations, a cell viability assay was performed.
After incubation, PrestoBlue (Thermo) viability reagent was added according to manufacturer’s instructions and samples were read microplate reader (BioTek Synergy HTX Multimode Reader, Agilent) [5]. Sample data was analyzed using Prism.
High Performance Liquid Chromatography Analysis.
To determine the incorporation rate of increasing concentrations of 6SG, HPLC analysis was performed according to previous methods [6][7].
After incubation, TRIzol (Thermo) reagent was added to each sample and processed according to manufacturer’s instructions [8]. Briefly, samples were homogenized, following which the aqueous phase containing RNA was collected. The supernatant was then collected and had any liquid removed, and then re-dissolved to be nano dropped (Thermo Scientific NanoDrop 2000 Spectrophotmeter) to find the RNA concentration in ng/uL.
Following RNA extraction, RNA was enzymatically digested to prevent oxidation of the sulfur group. Samples were prepared for HPLC and separated on a Supelco Discovery c18 (bonded phase silica 5 μM particle, 250’ 4.6mm) reverse phase column (Bellafonte PA, USA). HPLC buffers were 0.1 M TEAA in 3% acetonitrile (A) and 90% acetonitrile in water (B). The gradient was isocratic 0% B for 15 min, 0% to 10% B for 20 min, 10% to 100% B for 30 min, and a 5 min 100% B wash applied between runs to clean the HPLC column. Chromatograms were collected using Shimadzu software (Shimadzu CLASS-VP).
RESULTS.
Cell Viability Results.
After plate reading, data was plotted displaying concentration of 6SG treatment against relative fluorescence unit (Figure 2). PrestoBlue viability reagent fluoresces with metabolic activity, so relative fluorescence served as a proxy for cell viability in this assay. An increasing concentration of 6SG led to decreased cell viability, with higher concentrations approaching the cell viability of samples treated with positive control.

HPLC Analysis Results.
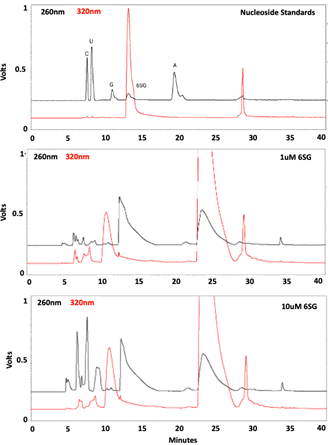
HPLC chromatograms were collected and formatted. There is a discrepancy between the peaks in the standards and what should have appeared in the experimental sample chromatograms.
DISCUSSION.
Cell viability assay results suggest that the ideal treatment of 6SG is 10 μM because this is the greatest concentration tested with limited cell death (Fig 2). Further testing may be performed to examine multiple trials of this concentration or concentrations ranging around this value.
HPLC results were found to be inconclusive because of their irregularity in comparison with the standards. The chromatogram of standards to which experimental samples were compared showed five peaks, one for each nucleoside standard and another for 6SG (Fig 3). However, when chromatograms of the experimental samples were collected, they displayed too many peaks which did not align with the standards in size or retention time (Fig 3). This could have been due to improper enzymatic digestion or contaminants may have been introduced during RNA isolation or digestion. Further testing needs to be performed to gather viable results for HPLC analysis.
CONCLUSION.
This research was conducted as part of a lab internship experience, creating inherent time limits on lab work. Being in the lab only on certain days as a student limited some lab work, which if not present could have allowed for another round of HPLC to gather conclusive results.
In the future HPLC methods need to be repeated to gain insight on 6SG’s incorporation rate into IFIT1 RNA. Following these experiments and finding a concentration of interest that balances cell viability and incorporation, a range of concentrations based on this initial research can be tested for the same results to find a 6SG concentration most adequate for PAR-dCLIP.
PTGR is vital to the immune systems, and the lab’s evidence suggests that IFIT1 plays a role in cellular gene expression. The next step is to characterize IFIT1’s functional importance. The research in this paper could help aid further characterization of IFIT1 using 6SG which would allow more specific identification of RNA transcripts. As we learn more about IFIT1 and other RBPs important to the immune system, we learn more about possible medical applications and treatments that could work with the immune system against pathogens.
This research has begun the process of supplementing work both in characterizing IFIT1 and the method of PAR-dCLIP at large through the utilization of 6SG. By adding to the PAR-dCLIP method, this research could be used to characterize other cap-and 5’ UTR-bound RBPs.
ACKNOWLEDGMENTS.
Thanks to all The Ascano Laboratory, especially Samantha Lisy and Dr. Manuel Ascano, and the SSMV faculty, especially Dr. Rebekah Stanton.
REFERENCES
- S. Lisy, K. Rothamel, M. Ascano, RNA Binding Proteins as Pioneer Determinants of Infection: Protective, Proviral, or Both? Viruses. 13, 2172 (2021).
- Y. M. Abbas, B. T. Laudenbach, S. Martínez-Montero, R. Cencic, M. Habjan, A. Pichlmair, M. J. Damha, J. Pelletier, B. Nagar, Structure of human IFIT1 with capped RNA reveals adaptable mRNA binding and mechanisms for sensing N1 and N2 ribose 2′-O methylations. Proceedings of the National Academy of Sciences. 114 (2017).
- C. Danan, S. Manickavel, M. Hafner, PAR-CLIP: A Method for Transcriptome-Wide Identification of RNA Binding Protein Interaction Sites. Methods in molecular biology (Clifton, N.J.). 1358, 153–173 (2016).
- K. Rothamel, S. Arcos, B. Kim, C. Reasoner, S. M. Lisy, N. Mukherjee, M. Ascano, ELAVL1 primarily couples mRNA stability with the 3′ UTRs of interferon-stimulated genes. Cell Reports. 35, 109178–109178 (2021).
- PrestoBlue Cell Viability Reagent for Microplates Protocol – US. www.thermofisher.com (2023), (available at https://www.thermofisher.com/us/en/home/references/protocols/cell-and-tissue-analysis/protocols/prestoblue-cell-viability-reagent-for-microplates-protocol.html).
- M. Hafner, M. Landthaler, L. Burger, M. Khorshid, J. Hausser, P. Berninger, A. Rothballer, M. Ascano, A.-C. Jungkamp, M. Munschauer, A. Ulrich, G. S. Wardle, S. Dewell, M. Zavolan, T. Tuschl, Transcriptome-wide Identification of RNA-Binding Protein and MicroRNA Target Sites by PAR-CLIP. Cell. 141, 129–141 (2010).
- A. Andrus, R. G. Kuimelis, Overview of Purification and Analysis of Synthetic Nucleic Acids. Current Protocols in Nucleic Acid Chemistry. 1 (2000).
Posted by buchanle on Tuesday, April 30, 2024 in May 2024.
Tags: 6SG, IFIT1, PAR-dCLIP, RNA Binding Protein

