Effect of Antisense Oligonucleotides on the Expression Level of Human Progranulin Associated with Frontotemporal Dementia
ABSTRACT
Progranulin deficiency is a major cause of frontotemporal dementia (FTD), a debilitating neurodegenerative disease. Granulin precursor (GRN) gene encodes progranulin protein (PGRN). GRN mRNA is targeted by microRNA-29b (miR-29b) which decreases GRN expression, resulting in PGRN insufficiency. This project focused on using antisense oligonucleotides (ASOs) to increase PGRN levels by adding ASOs targeting the miR-29b binding site to H4 neuroglioma cells. It was found that the ASOs significantly increased the amount of PGRN. Additionally, this study examined if the ASOs only act through the miR-29b binding site. It was determined that combining the miR-29b inhibitors with the ASOs resulted in no significant increase in PGRN levels, thus implying that the ASOs only work through the miR-29b binding site. These findings suggest that using the ASOs to compete for the miR-29b’s binding site could stop the molecule from inhibiting PGRN expression and ultimately lead to higher levels of PGRN in the brain, which would treat progranulin-deficient FTD.
INTRODUCTION.
Frontotemporal dementia (FTD) is a neurodegenerative disease caused by the loss of nerve cells in the brain’s frontal and temporal lobes [1,2]. It is the second most common form of neurodegenerative dementia, making up 25% of all instances of dementia in patients ages 65 and older [3]. The disease is characterized by abnormal changes in behavior, difficulty with language such as speech impairments, and movement disorders. Although FTD is a debilitating and degenerative disease, a definitive treatment or cure has yet to be discovered. However, recent advancements suggest that a treatment could be achieved by targeting the protein progranulin (PGRN), which is expressed through the granulin precursor (GRN) gene [4,5]. Patients suffering from FTD exhibit mutations on the GRN which have been reported to result in haploinsufficiency [6]. Based on such information, it is logical to devise a treatment for FTD by increasing the expression of progranulin from the remaining wild-type GRN allele. MicroRNAs are short non-coding RNAs that regulate gene expression. They regulate protein levels of about one third of all proteins in humans [7] by generally binding to the 3′ UTR (untranslated region) of their target mRNAs and repressing translation or decreasing the stability of their target mRNAs [8]. MicroRNA-29b (miR-29b) has been reported to lower PGRN levels by binding to the miR-29b binding site of the human GRN mRNA [9]. A treatment modality that has recently been receiving much attention is antisense oligonucleotides (ASOs), which are single-stranded pieces of DNA whose structure allows for complementary base-pairing with a target mRNA [10]. The ASOs would then theoretically compete with the microRNA for the binding site, preventing the inhibition of the GRN mRNA, therefore increasing the amount of PGRN and prohibiting the progression of FTD.
In this study, two ASOs, differing in their binding site on the 3’ UTR, were designed to target the miR-29b binding site of the GRN mRNA. Both ASOs were found to have blocked miR-29b’s targeting sequence, preventing the microRNA and mRNA from interacting and thus increasing PGRN levels. However, in order to determine if the ASOs would out-compete the miR-29b, it was necessary to demonstrate that its binding site was the only site that the ASOs could work through despite the presence of other microRNA binding sites, such as miR-659 [11] and miR-107 [12], on the GRN gene. This was another aim of the experiment. To study this, miR-29b inhibitors were added along with the ASOs, and the progranulin levels before and after adding the ASOs were measured using an ELISA assay (enzyme-linked immunoassay).
MATERIALS AND METHODS.
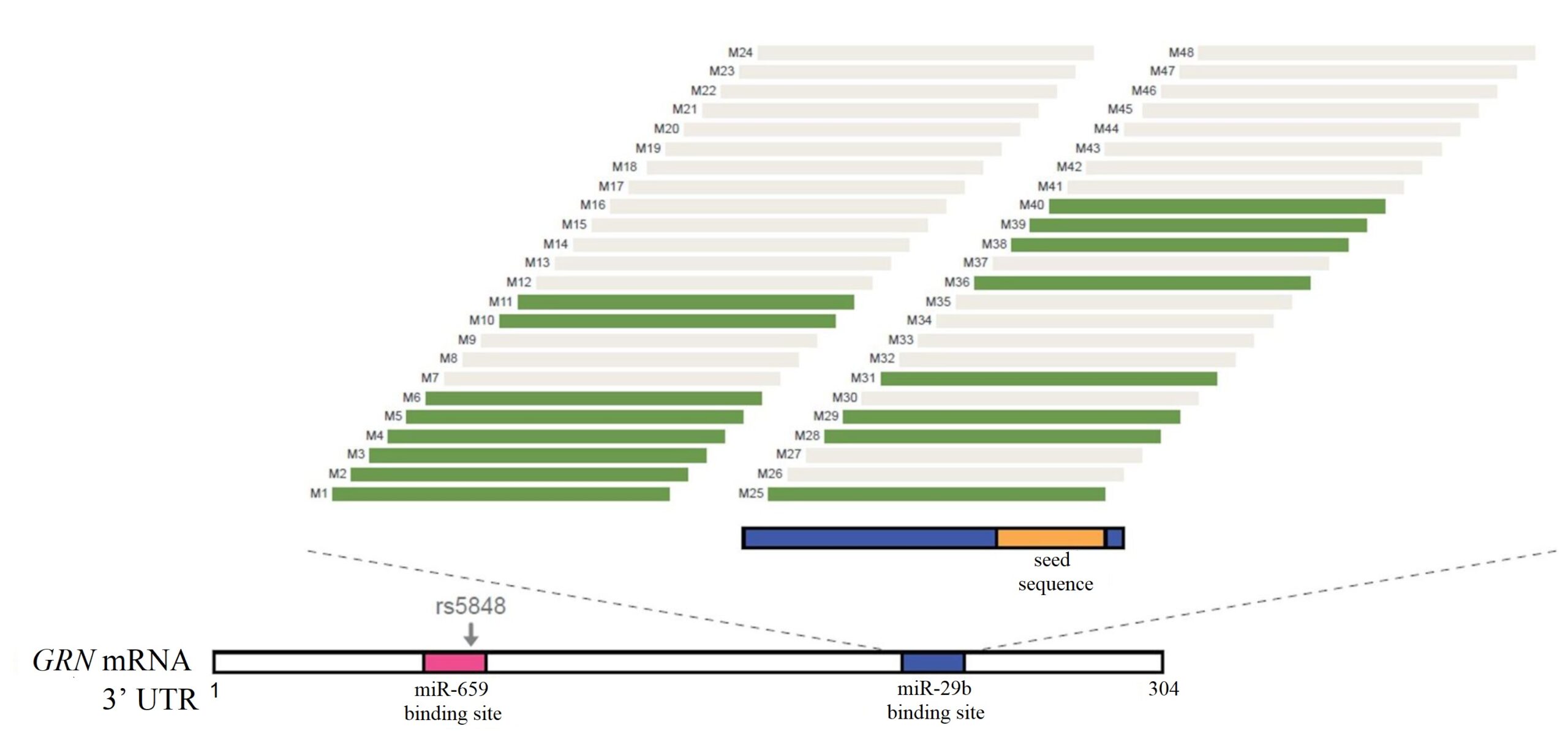
48 ASOs (Ionis Pharmaceuticals) targeting the miR-29b binding site were created (referred to as M1 – M48). Initial screens identified 16 ASOs could potentially increase PGRN levels, as shown in green in Figure 1. M5 and M10, referred to as ASO1 and ASO2 respectively, were used in this experiment.
Figure 1. Schematic representation of the areas on 3’ UTR that the ASOs bind to.
The sequences of the ASOs used in this experiment were:
ASO1: ACTGAAACGGGGAGGGGA
ASO2: GGTCCACTGAAACGGGGA
Control ASO: TTAGTTTAATCACGCTCG
Control ASO was created using the same method as progranulin targeting ASOs, only with a scrambled sequence.
ASOs Increase PGRN Levels.
To examine the effects of the ASOs on PGRN levels, H4 neuroglioma (American Type Culture Collection HTB-148) cells were cultured, plated, and treated with 20 μM of ASO1, ASO2, or control ASOs. Next, an ELISA assay was performed.
- Culture cells
H4 neuroglioma cells were used to simulate a human brain affected with a neurological disorder [13]. The cells were suspended in Dulbecco’s Modified Eagle Medium (DMEM) culture medium and centrifuged at 125 x g. The cell pellet was then re-suspended in DMEM medium. The cells were finally incubated at 37°C in a 5% CO2 incubator.
- Plate cells
The neuroglioma cells were washed with PBS 1X, trypsinized using 0.05% trypsin EDTA, and placed at 37°C in a 5% CO2 incubator for 3 minutes. DMEM (with added antibiotics penicillin and streptomycin) was then added to the dish to terminate the trypsinization. Next, the detached cells were washed with DMEM and centrifuged at 0.3 x g. The pellet was then re-suspended in DMEM and read on a cell counter (Cyto Cell Counter), after which the cells were pipetted into the wells of the plate and incubated for 24 hours at 37°C in a 5% CO2 incubator.
- Treat cells
DMEM was added to ASO1, ASO2, or control ASOs to make the final concentration 20 μM. After aspirating the media from the wells, ASOs were added into each of the wells and the plate was placed at 37°C in a 5% CO2 incubator for 24 hours.
- ELISA (ELISA kit, R&D Systems)
ELISA kit instructions were followed.
Determine If ASOs Act through miR-29b.
In order to determine if the ASOs only act through the miR-29b binding site, different concentrations of the miR-29b inhibitor (0 µM, 20 µM, 40 µM, 70 µM, and 100 µM, Ionis Pharmaceuticals) were tested against 20 µM each of ASO1, ASO2, or control ASOs. Cells were cultured, plated, and treated the same way as described above. An ELISA assay was then conducted to measure the amount of PGRN corresponding to the various amounts of inhibitor added following the aforementioned steps.
Statistical Analysis.
t-tests were conducted on the results of testing the ASOs on increasing PGRN levels. Two-way ANOVA followed by Bonferroni’s post hoc tests were conducted on the results of testing if the ASOs act through the miR-29b binding site. P ≤ 0.05 was considered statistically significant.
P-value symbols and meanings shown in the figures are:
ns P > 0.05
* P ≤ 0.05
** P ≤ 0.01
*** P ≤ 0.001
**** P ≤ 0.0001
All tests were performed in triplicates with the exception of analyzing the effect of ASO1 on PGRN levels which was done 11 times. The data was presented as means -/+ SD.
RESULTS.
ASOs Increase PGRN Levels.
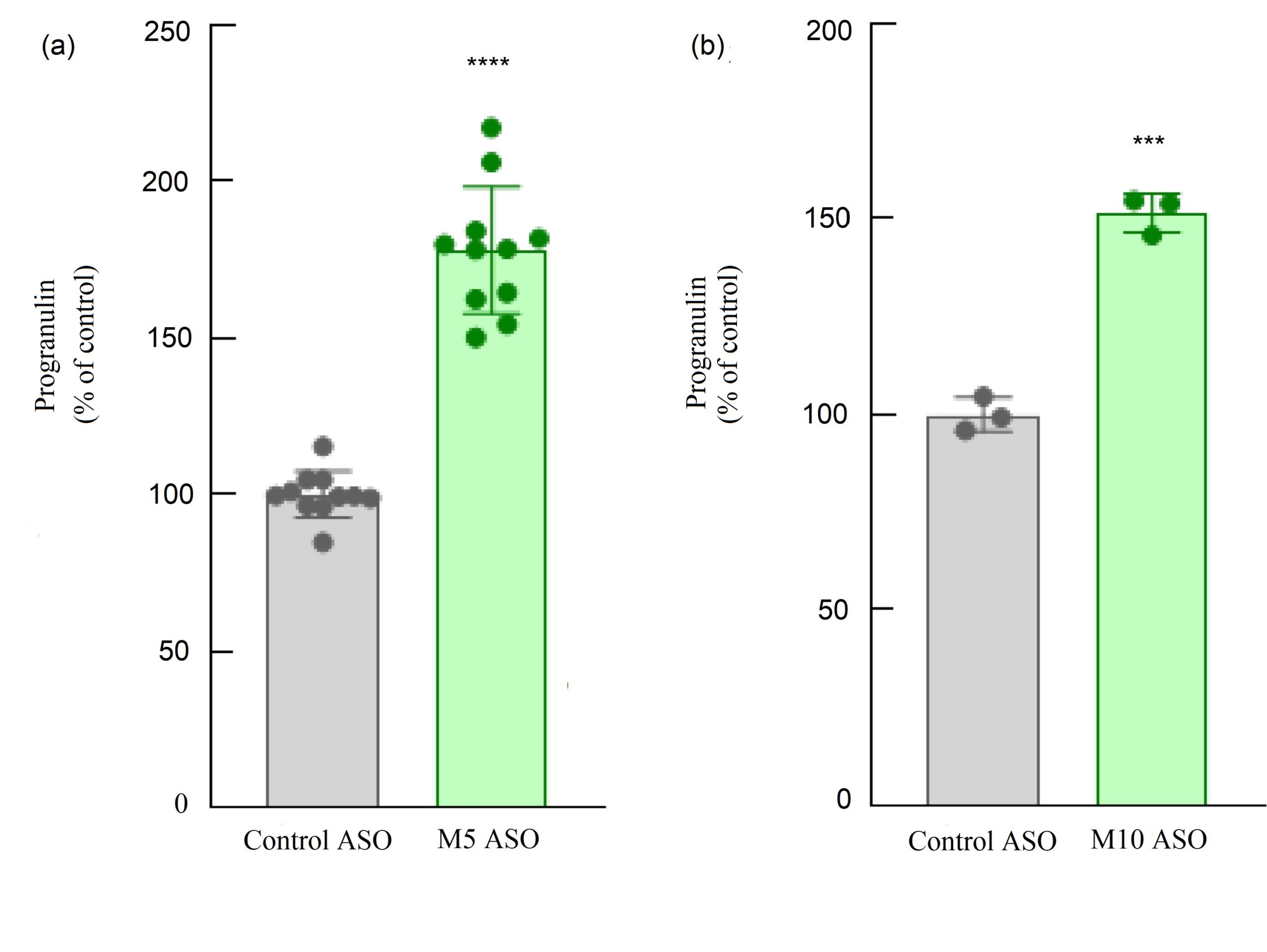
Figure 2. H4 neuroglioma cells were treated with ASO1, ASO2 or control ASOs. The relative progranulin level of the control was set as 100%. Adding ASO1 and ASO2 caused a significant increase in progranulin concentration compared with control.
PGRN levels after adding the ASOs were studied, and the t-test indicated that both ASOs caused a significant increase in PGRN levels.
As indicated in Figure 2 (a), when adding ASO1, there was a significant increase in PGRN levels compared with control (p < 0.0001). This finding was supported in all 11 trials of this test. These results were mirrored in the experiment with ASO2 as shown in Figure 2 (b), where all three trials showed that adding ASO2 caused a significant increase in PGRN level (p < 0.001).
Determine If ASOs Act through miR-29b.
In order to determine if the ASOs act only through the miR-29b binding site, the miR-29b inhibitor was added to block the miR-29b binding site and prevent the ASOs from binding to the same site and increasing PGRN levels. A combination of increasing concentrations of the miR-29b inhibitor was added to the cells at the same time along a stable concentration of control, ASO1, or ASO2.
As shown in Figure 3 (a), when there was no inhibitor presented, adding ASO1 significantly increased PGRN concentration compared with the control ASOs. This was consistent with what we had observed above. However, when the miR-29b inhibitors were added, no matter what concentration was used, the effects of ASO1 were essentially nullified because neither one created an increase in PGRN levels compared to the control ASOs plus inhibitor.
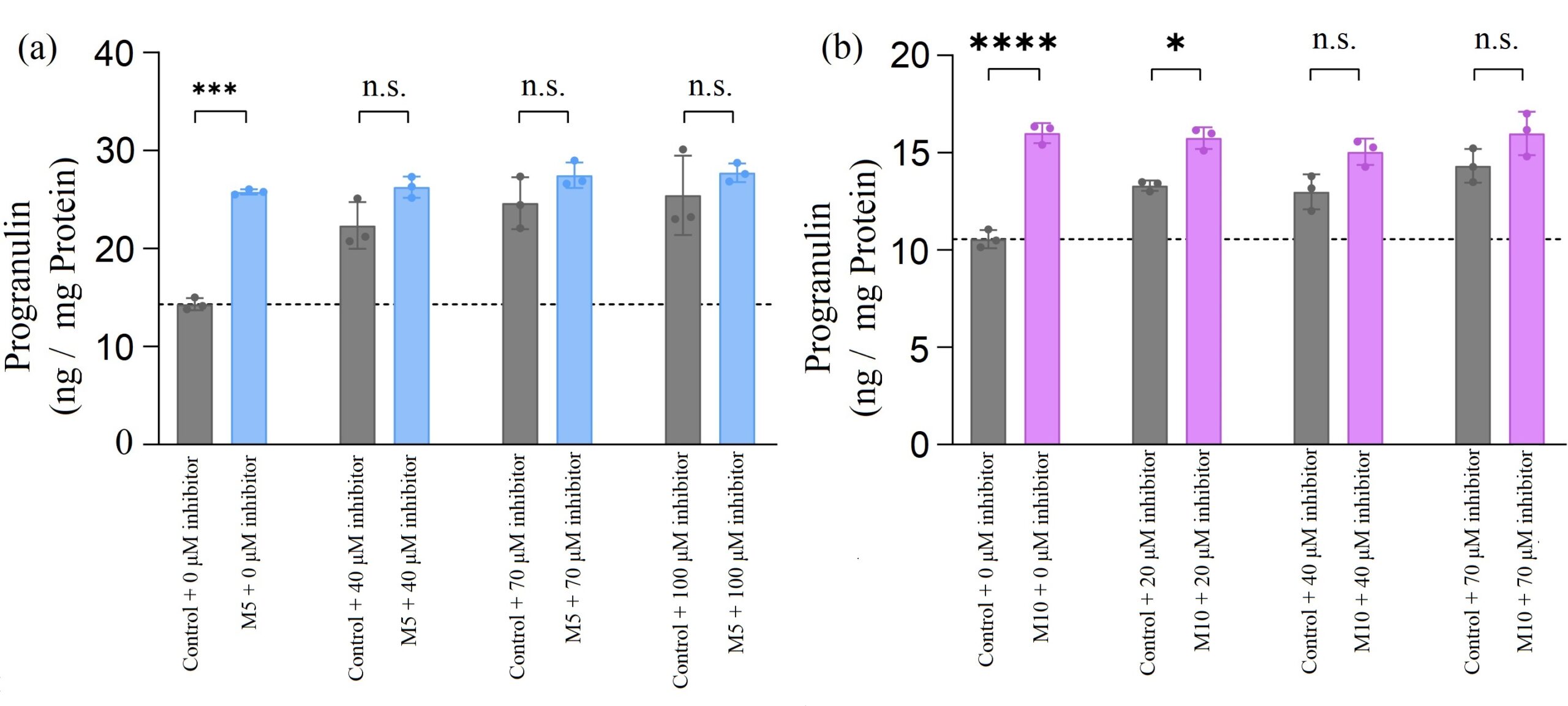
Figure 3. H4 neuroglioma cells were treated with increasing concentrations of miR-29b inhibitor and 20 µM of ASOs (ASO1, ASO2, or control ASO). (a) There was no significant change in progranulin level when cells were treated with ASO1 and inhibitor compared with control ASO and inhibitor. (b) When inhibitor concentration was greater than 40 µM, there was no significant change in progranulin level when cells were treated with ASO2 and inhibitor compared with control ASO and inhibitor.
In Figure 3 (b), when ASO2 was tested with a low concentration of the miR-29b inhibitors (20 µM), PGRN concentrations increased significantly compared with the control ASOs, as the binding sites had not been completely saturated with the inhibitor. Therefore, ASO2 was able to bind to the target site and increase PGRN level. However, when ASO2s were tested with the subsequent higher concentrations of the inhibitors, there was no significant change observed compared to the control ASOs.
DISCUSSION.
ASOs Increase PGRN Levels.
Adding both ASOs caused a significant increase in PGRN concentration. This increase is due to the binding of the ASOs to the miR-29b binding site, inhibiting the binding of miR-29b to the same site. As a result, PGRN levels were able to increase.
The increase in PGRN with ASO2 compared to the control ASO was less pronounced than it was with ASO1. There are multiple possibilities for this. First, there is an optimal position for each ASO in terms of their ability to sterically block the binding site of the miR-29b [14], which is why multiple ASOs were designed to span the entire binding site and screened in order to find which ASOs worked best. Second, biophysical properties of the ASOs, such as how tightly they bind to the GRN mRNA, also impact their efficiency [15]. Lastly, the secondary structure of the GRN mRNA may favor the binding of certain ASOs. mRNAs have a lot of self-interactions (forming stems and loops for example), so the ASOs targeting regions that are accessible for binding (such as in a big loop) are expected to have stronger effects [16].
Determine If ASOs Act through miR-29b.
When all the miR-29b binding sites were saturated with the miR-29b inhibitor, adding the ASOs did not significantly increase PGRN amount compared to the control ASOs.
These non-significant changes in PGRN levels can be explained by the lack of binding sites for the ASOs after being saturated by the inhibitor, thus limiting the effects of the ASOs. Had there been a separate binding site other than the miR-29b, a significant increase in PGRN concentration would have been observed, as the ASOs would have bound elsewhere.
Thus, Figures 3 demonstrate that ASOs only act through the miR-29b binding site.
CONCLUSION.
This study determined that ASOs can increase PGRN levels significantly while only working through the miR-29b binding site. These findings suggest that using ASOs could inhibit the decrease in PGRN caused by miR-29b, thus providing effective treatment for progranulin-deficient FTD. This could come in the form of increasing target protein levels through pharmacological means as well as creating ASO-based therapies for haploinsufficiency-related diseases.
To further demonstrate that ASOs work solely through the miR-29b binding site, a future study could use luciferase assays by mutating the miR-29b binding site to theoretically prevent the ASOs from binding. A preliminary test had been conducted as part of this project but generated inconsistent results which might have been due to not enough luciferase/renilla DNA being transfected into the cells and caused the low signal in the luciferase assays. Due to the low signal, there was relatively more noise in the results, which more than likely caused the inconsistent results. Therefore, for future luciferase assays, more DNA needs to be transfected into the cells in order to get a stronger signal.
Another possible future experiment could be trying to block other binding sites besides just the miR-29b. For instance, miR-659 and miR-107 are also promising choices since they also regulate progranulin expression. It could be possible to block more than one binding site and increase progranulin levels even further.
ACKNOWLEDGMENTS.
Sincere thanks to Dr. Andrew Nguyen for his guidance and support.
REFERENCES.
- J. Gass, A. Cannon, I. R. Mackenzie, B. Boeve, M. Baker, J. Adamson, R. Crook, S. Melquist, K. Kuntz, R. Petersen, K. Josephs, S. M. Pickering-Brown, N. Graff-Radford, R. Uitti, D. Dickson, Z. Wszolek, J. Gonzalez, T. G. Beach, E. Bigio, N. Johnson, S. Weintraub, M. Mesulam, C. L. White, B. Woodruff, R. Caselli, G. Y. Hsiung, H. Feldman, D. Knopman, M. Hutton, R. Rademakers, Mutations in progranulin are a major cause of ubiquitin-positive frontotemporal lobar degeneration. Hum. Mol. Genet. 15, 2988-3001 (2006).
- M. Cruts, I. Gijselinck, J. van der Zee, S. Engelborghs, H. Wils, D. Pirici, R. Rademakers, R. Vandenberghe, B. Dermaut, J. J. Martin, C. van Duijn, K. Peeters, R. Sciot, P. Santens, T. De Pooter, M. Mattheijssens, M. Van den Broeck, I. Cuijt, K. Vennekens, P. P. De Deyn, S. Kumar-Singh, C. Van Broeckhoven, Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 442, 920-924 (2006).
- J. J. Young, M. Lavakumar, D. Tampi, S. Balachandran, R. R. Tampi, Frontotemporal dementia: latest evidence and clinical implications. Therapeutic advances in psychopharmacology. 8 (1), 33–48 (2018).
- E. D. Huey, J. Grafman, E. M. Wassermann, P. Pietrini, M. C. Tierney, B. Ghetti, S. Spina, M. Baker, M. Hutton, J. W. Elder, S. L. Berger, K. A. Heflin, J. Hardy, P. Momeni, Characteristics of frontotemporal dementia patients with a progranulin mutation. Annals of neurology. 60 (3), 374–380 (2006).
- A. D. Nguyen, T. A. Nguyen, L. H. Martens, L. L. Mitic, R. V. Farese Jr., Progranulin: at the interface of neurodegenerative and metabolic diseases. Trends Endocrinol. Metab. 24 (12), 597-606 (2013).
- I. G. Cannell, Y.W. Kong, M. Bushell, How do microRNAs regulate gene expression? Biochem. Soc. Trans. 36 (Pt 6), 1224-1231 (2008).
- S. M. Hammond, An overview of microRNAs. Advanced drug delivery reviews. 87, 3-14 (2015).
- J. Terryn, C. M. Verfaillie, P. Van Damme, Tweaking progranulin expression: therapeutic avenues and opportunities. Front. Mol. Neurosci. 14, 713031 (2021).
- J. Jiao, L. D. Herl, R. V. Farese Jr, F. B. Gao, MicroRNA-29b regulates the expression level of human progranulin, a secreted glycoprotein implicated in frontotemporal dementia. PLOS ONE. 5 (5), e10551 (2010).
- K. M. Schoch, T. M. Miller, Antisense oligonucleotides: translation from mouse models to human neurodegenerative diseases. Neuron. 94 (6), 1056-1070 (2017).
- R. Rademakers, J. L. Eriksen, M. Baker, T. Robinson, Z. Ahmed, S. J. Lincoln, N. Finch, N. J. Rutherford, R. J. Crook, K. A. Josephs, B. F. Boeve, D. S. Knopman, R. C. Petersen, J. E. Parisi, R. J. Caselli, Z. K. Wszolek, R. J. Uitti, H. Feldman, M. L. Hutton, I. R. Mackenzie, N. R. Graff-Radford, D. W. Dickson, Common variation in the miR-659 binding-site of GRN is a major risk factor for TDP43-positive frontotemporal dementia. Human Molecular Genetics. 17, 3631–3642 (2008).
- W. X. Wang, B. R. Wilfred, S. K. Madathil, G. Tang, Y. Hu, J. Dimayuga, A. J. Stromberg, Q. Huang, K. E. Saatman, P. T. Nelson, miR-107 regulates granulin/progranulin with implications for traumatic brain injury and neurodegenerative disease. The American journal of pathology. 177 (1), 334–345 (2010).
- A. Slanzi, G. Iannoto, B. Rossi, E. Zenaro, G. Constantin, In vitro models of neurodegenerative diseases. Front. Cell Dev. Biol. 8, 328 (2020).
- R. Kole, A. Krainer, S. Altman, RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov. 11, 125–140 (2012).
- K. H. Altmann, D. Fabbro, N. M. Dean, T. Geiger, B. P. Monia, M. Müllert, P. Nicklin, Second-generation antisense oligonucleotides: structure-activity relationships and the design of improved signal-transduction inhibitors. Biochem Soc Trans. 24 (3), 630–637 (1996).
- N. Dias, C. A. Stein, Antisense oligonucleotides: basic concepts and mechanisms. Mol. Cancer. Ther. 1 (5), 347-355 (2002).
Posted by John Lee on Saturday, May 7, 2022 in May 2022.
Tags: Antisense Oligonucleotides, FTD, miR-29b, Progranulin