Confirming the Suitability of Unnatural Amino Acids as Labels to Monitor Structural Changes in Amyloid-Beta(40) with Two-Dimensional Infrared Spectroscopy
ABSTRACT
Alzheimer’s disease (AD) is a neurodegenerative disease that affects 1 in 9 Americans over age 65. AD is often characterized by the presence of plaques containing amyloid-beta (Aβ) fibrillar aggregates. The pathways by which Aβ monomers misfold and aggregate into fibrils are not well understood and are crucial to understanding how it becomes harmful. This study employed two-dimensional infrared spectroscopy (2D IR) to monitor changes in peptide structure throughout aggregation. We incorporated azidohomoalanine (AHA) at the methionine-35 position within Aβ(40) as to serve as a site-specific vibrational probe. The azido vibrational mode falls within the cell-silent region of infrared spectra, allowing us to examine its specific behavior without interference from water or standard peptide vibrational modes. Here, we report preliminary 2D IR spectra of AHA-labeled Aβ(40). We induce aggregation by lowering the pH from 8.5 to 7.5 and observe a clear frequency shift of the AHA label from 2112 cm-1 to 2104 cm-1. These findings indicate that the AHA label is a sensitive probe of Aβ structure and can be used in the future to track structural changes throughout the aggregation process, even in complex biological media.
INTRODUCTION.
Alzheimer’s Disease (AD) is a very common neurodegenerative disease in seniors that affects 1 in 9 people over the age of 65 in the United States [1]. AD causes memory loss and decline in cognitive function. Its progression is often associated with the presence of amyloid-beta (Aβ) oligomers, a toxic species preceding the formation of fibrils, in the brain [2, 10]. Aβ(42) and Aβ(40) are both naturally occurring isoforms of Aβ in humans, differing only by two residues at the C-terminus. While Aβ(42) is more indicative of AD [3], both forms are implicated in AD and here we choose to focus on Aβ(40) which is known to be easier to work with for biophysical studies. The understanding of the aggregation of Aβ(40) from monomer to oligomer and fibril formations could be crucial to understanding the underlying causes of the disease, but as of yet little is known about the aggregation pathways of Aβ(40) [4].
Two-dimensional infrared spectroscopy (2D IR) is a relatively new method to analyze protein structure and dynamics [5, 6]. Because of its sensitivity to individual molecular bonds, isotopes and unnatural amino acids (UAAs) containing unique functional groups can be used to “label” specific positions along the peptide chains and thus capture the evolution of structure at individual amino acids. [5-7]. Traditionally, 2D IR studies of proteins have focused on the amide I’ spectra region (1600 to 1700 cm-1), where vibrations of the amide groups of the peptide backbone are highly sensitive to changes in protein structure. To gain single-residue structural resolution, individual amino acids can be isotopically labeled, but both the native amide I’ and isotope-labeled signals can be obscured by other vibrations within the peptides and by water itself. To reduce such interference, most infrared studies of proteins must be performed using isolated samples in deuterated buffers. The UAA azidohomoalanine (AHA) has a characteristic vibration of the azido functional group within the “cell-silent” region of the infrared spectrum (1800-2700 cm-1), where water and peptide vibrational modes do not create interfering signals. Thus, AHA provides a unique approach to probing protein structure via its azido vibration as opposed to the native amide I’ vibrations of the peptides [5, 9].
The goal of this project was to determine whether AHA can be used as a UAA IR label to report more specific structural and kinetic information about the aggregation process of Aβ(40). In this study, AHA replaces methionine-35 (M35) because of their similar structures [7]. We expected to find that using labeled Aβ(40) would allow us to see clear signal from the peptide without congestion from other vibrations. This project also worked towards streamlining the synthesis, purification, disaggregation, pH, buffer, and temperature parameters to obtain reproducible results.
MATERIALS AND METHODS.
Sample Preparation. AHA-labeled Aβ(40) was prepared using microwave-assisted solid phase peptide synthesis on a Liberty Blue (CEM) peptide synthesizer. Residue M(35) was replaced with an AHA label. Synthesized Aβ(40) was cleaved from the resin using a solution of 90% trifluoroacetic acid, 5% 1-2 ethanedithiol, 2.5% anisole, and 2.5% thioanisole. The peptide was filtered through the solution several times to remove the resin.
The crude peptide was purified using size exclusion chromatography (SEC) using Superdex 75 Increase 10/300 GL SEC column with ATKA Pure Fast Protein Liquid Chromatography (FLPC). Tris(hydroxymethyl)aminomethane (TRIS) was used as a running buffer and the peak corresponding to the monomer peptide was fraction collected in between running the program. The collected peptide was immediately desalted to fully purify it. A Waters HLB 3 cc/60 mg cartridge and Waters extraction module were used to separate the peptide from the TRIS buffer.
Matrix-Assisted Laser Desorption/Ionization (MALDI) mass spectrometry was used to confirm successful synthesis of Aβ(40)-AHA.
2DIR Methods. The 2D IR instrumentation is described in detail in Weeks et al., 2022 [8]. Data was plotted and analyzed using custom MATLAB scripts. 2D IR contour plots show the intensity of infrared vibrational signals as a function of pump and probe frequency.
An important goal of this project was to ensure we could see signal in 2DIR from the AHA label and to determine whether the AHA peak changed as protein structure changed. To do this, the concentration and pH of Aβ(40)-AHA in a 20 mM TRIS buffer were adjusted and the samples scanned using 2DIR spectroscopy in the spectral region ranging from 2000 to 2400 cm-1.
After demonstrating that AHA does show a signal in 2DIR and finding optimal conditions for disordered and aggregate form, the next step was to find the ideal other starting conditions. Specifically, those conditions were time after SEC purification for ideal stability, whether to use hexafluoroisopropanol (HFIP) or NH4OH for disaggregation, ideal concentration, buffer type, pH, and temperature. pH and temperature tests were also conducted on unlabeled Aβ(40) to find their effects on aggregation. In those tests, Aβ(40) was stored in buffers with pH ranging from 7.4-8.0 and at either room temperature or 37℃. After finding the ideal conditions, the next step is a kinetic run at those conditions with AHA. These steps are still in process.
RESULTS.
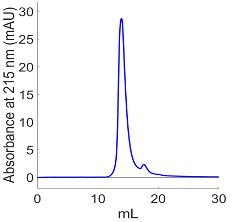
SEC is a method of purification in which molecules are separated by their size, with smaller particles coming out later. In SEC purification of Aβ(40)-AHA, a large peak (Fig. 1) shows that most of the peptide is coming off at one size, which suggests that the purified peptide is likely monomeric as aggregated peptides would likely form a wide range of oligomeric and fibrillar species. The small peak to the right of the main peak indicates that another conformation may be present, which is not ideal but based on intensities this species is only a minor contributor.
Figure 1. SEC indicates that we are collecting our peptide, but also the presence of a possible aggregate form or other larger molecule.
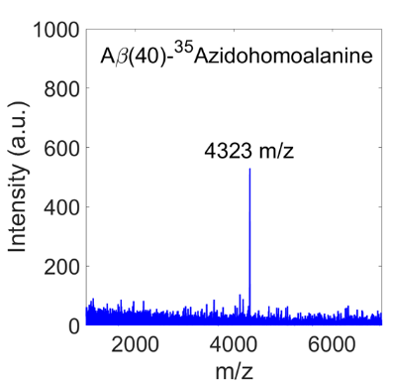
The data from MALDI confirms that we do have the correct peptide (Fig. 2). The peak at a mass-to-charge ratio of 4323 m/z corresponds to a molecular weight of 4323 g/mol with a charge of 1, which matches the molecular weight of Aβ(40)-AHA.
Figure 2. The peak from MALDI indicates that we have the correct peptide, because the mass-to-charge ratio matches the molecular weight of Aβ(40)-AHA
Regarding conditions for aggregation, the pH tests conducted on unlabeled Aβ(40) showed us that a higher pH of 8.5 should maintain the Aβ(40) in disordered form, and a lower pH of 7.5 should aggregate the Aβ(40) into oligomer or fibril form. These results were confirmed by 2D IR spectra of the backbone amide I’ vibrational modes (spectra not shown). Of the subsequent tests on Aβ(40)-AHA, the conditions that have so far been tested that have led to aggregation have been 1 week of time after SEC, disaggregation by HFIP, 10 or 20 mM Phosphate as a buffer, 7.6 buffer pH, and 37°C.
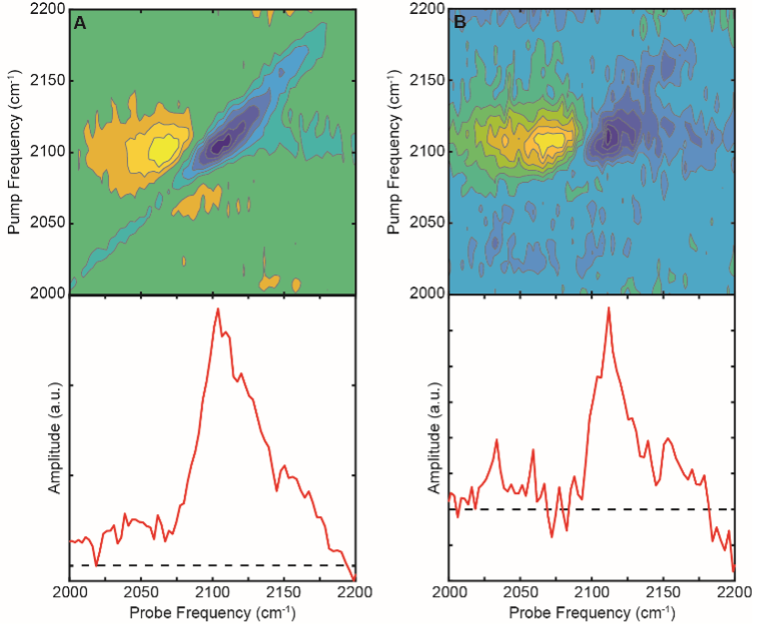
Preliminary results from 2D IR spectroscopy indicate that we can see signal from the azido group of Aβ(40)-AHA within the cell-silent region. After running many scans, the sample showed signal both at 3.29 mM Aβ(40)-AHA in 20 mM Tris buffer with pH~7.5 and 2.5 mM Aβ(40)-AHA in 20 mM Tris at pH 8.5 (Figure 3). In the literature, UAAs are generally studied between 5 and 100 mM [4]. These spectra thus represent among the best sensitivity in the field.
Figure 3. 2D IR spectra (top) and diagonal “slices” (bottom) of Aβ(40)-AHA at pH 7.5 (A) and pH 8.5 (B). At pH 7.5, Aβ(40)-AHA is aggregated and shows a peak centered around 2104 cm-1. At pH 8.5, Aβ(40)-AHA remains mostly disordered/monomeric and displays a peak around 2112 cm-1
Differences in the signal such as the frequencies of peaks and the appearance of cross peaks are expected to indicate structural alterations when Aβ(40)-AHA is introduced to different buffer conditions. Cell-silent region data from labeled peptides are shown in Figure 3. At pH 8.5, which maintains the disordered structure of the peptide, we can see a peak centered around 2112 cm-1(shown in Figure 3B). At pH 7.5, which leads to rapid aggregation into amyloid fibrils, a peak around 2104 cm-1 appears (shown in Figure 3A).
DISCUSSION.
We have found that AHA can serve as a structural probe for 2D IR studies of Aβ(40). When Aβ(40)-AHA is disordered and monomeric, the azido group of AHA produces a peak centered around 2112 cm-1, whereas aggregated Aβ(40)-AHA displays a peak centered at 2104 cm-1. Moving forward, such frequency shifts can be analyzed to determine the pathways of oligomerization relevant to AD.
Furthermore, we have identified sample conditions that lead to immediate aggregation. This is critical knowledge for future studies aiming to use 2DIR to analyze protein aggregation pathways as we will need to be able to track protein structural changes in real time. Thus, conditions that produce aggregates more quickly than the timescale of our measurements must be avoided.
Although we have demonstrated that AHA can serve as a sensitive probe of protein secondary structure, we need to confirm that the AHA label does not alter the kinetic process of labeled variant compared to the wild type. As native Aβ(40) does not contain any azido vibrations, spectral signatures in the amide I’ region must be compared between wild type Aβ(40) and AHA-labeled Aβ(40) variants in order to detect any differences in aggregation rates or peptide backbone structure. Other UAA, like isocyanoalanine and isocyanotryptophan, will be explored for sidechain labeling in order to determine which sidechain modifications are least disruptive based on the position at which they are inserted [4]. Once appropriate UAA labels have been identified as not altering the aggregation process, we can continuously collect 2D IR data throughout the aggregation of Aβ(40) and analyze the spectral signatures of the labels to better understand the mechanisms by which Aβ(40) aggregates.
In the future, we can continue this project by analyzing AHA’s specific behavior in different conditions that are relevant to AD. Such biologically relevant conditions would not be accessible for 2D IR studies due to interfering signals from other biomolecules and water. Ultimately, these studies will give us unique insight on how Aβ(40) behaves in a more realistic environment, which is a long-term goal of this project.
ACKNOWLEDGMENTS.
The Buchanan Research Group all helped guide me through the process. The School for Science and Math at Vanderbilt (SSMV) provided me with the opportunity to do this internship. Dr. Menton Deweese was my advisor for project from the SSMV.
REFERENCES
- Alzheimer’s Disease and Dementia, Alzheimer’s disease facts and figures
- R. Sarroukh, et al., ATR-FTIR: A ‘rejuvenated’ tool to investigate amyloid proteins, BBA- Biomembranes, 1828, 2328–2338 (2013)
- S. G. Younkin, Evidence that Aβ42 is the real culprit in Alzheimer’s disease, Ann. Neurol., 37, 287–288 (1995)
- D. M. Walsh, D. J. Selkoe, Amyloid β-protein and beyond: The path forward in Alzheimer’s disease, Curr. Opin. Neurobiol., 61, 116–124 (2020)
- M. C. Thielges, Transparent window 2D IR spectroscopy of proteins, J. Chem, Phys., 155, 040903 (2021)
- A. Ghosh, et al., Watching proteins wiggle: Mapping structures with two-dimensional infrared spectroscopy, Chem. Rev., 117, 10726–10759 (2017)
- R. Bloem, et al., Ligand binding studied by 2D IR spectroscopy using the Azidohomoalanine label, J. Phys. Chem B, 116, 13705–13712 (2012)
- W. B. Weeks, C. J. Tainter, and L. E. Buchanan, Investigating the effects of N-terminal acetylation on KFE8 self-assembly with 2D IR spectroscopy, Biophysical Journal, 121, 1549–1559 (2022)
- C. Zanobini, et al., Azidohomoalanine: A minimally invasive, versatile, and sensitive infrared label in proteins to study ligand binding, J. Phys. Chem. B, 122, 10118–10125 (2018)
- W. Klein, Targeting small AB oligomers: The solution to an Alzheimer’s disease conundrum? TINS, 24, 219-224 (2001)
- J. K. Kasim, I. Kavianinia, J. Ng, P. W. Harris, N. P. Birch, and M. A. Brimble, “Efficient synthesis and characterisation of the amyloid beta peptide, AΒ1–42, using a double linker system,” Organic & Biomolecular Chemistry, 17, 30–34 (2019)
Posted by John Lee on Tuesday, May 30, 2023 in May 2023.
Tags: Amyloid-Beta, label, oligomer