Computation simulations of IL-13Rα2 receptor+ CAR T-cell targeting high-grade glioma
ABSTRACT
Gliomas are aggressive tumors arising from the uncontrolled proliferation of glial cells, often leading to severe neurological complications. Despite advancements in treatment, gliomas remain challenging due to their invasive nature and resistance to conventional therapies. CAR T-cell therapy, which involves genetically modifying a patient’s T cells to target specific cancer markers, has emerged as a promising strategy for glioma treatment. However, optimizing receptor-target interactions is crucial to enhancing therapeutic efficacy. We hypothesize that the CD45 and CD62 receptors (present on the surface of CAR T cells) bind to the specific binding site on the surface of the IL-13Rα2 receptor, resulting in their interactions. In this study, we investigate the molecular interactions between CD45, CD62, and IL-13Rα2, a glioma-associated receptor, to improve CAR T-cell targeting. The receptor structures were predicted using AlphaFold 3, and binding sites were identified using the Graph Attention Site Prediction (GrASP) method. Molecular docking simulations were conducted using the HDOCK server to analyze CD45-IL-13Rα2 and CD62-IL-13Rα2 interactions. Our results confirm that the predicted binding sites align with the docking interactions, validating the reliability of graph neural network-based site predictions. These findings provide critical insights into receptor binding mechanisms, paving the way for more precise CAR T-cell designs. By enhancing CAR T-cell targeting strategies, this research contributes to the development of more effective immunotherapies for high-grade gliomas, potentially improving patient outcomes.
INTRODUCTION.
Glioma is a cancerous brain/spine tumor arising from glial cells(1). In the United States, there are 6 cases of glioma diagnosed per 100,000 individuals every year. Gliomas are one of the most prevalent and hard-to-tame diseases(1). This is due to extreme difficulty in treatment and limited options for treatment(2). Currently, the main treatment methods include surgery, radiation therapy, chemotherapy, and immunotherapy(3). Glioma treatments often cause side effects like incomplete removal, damage to healthy brain tissue, and treatment resistance (4). Gliomas have a high recurrence rate, with 50-90% of cases recurring within 6–12 months after initial treatment, depending on the grade and type of glioma(5).
Targeting and killing cancer cells effectively has been a problem for ages, but there may be a solution(5). The T-cell can hunt down and destroy cancer cells by genetically modifying a patient’s T-cells, a type of white blood cell, and producing chimeric antigen receptors (CARs)(6). This is known as CAR T-cell therapy but does not come without its risks and side effects, such as neurological problems and cytokine release syndrome(5). Furthermore, CAR T-cell therapy only kills certain types of cancers and lasts for a short duration(7).The development of CAR-T cell therapy is a promising treatment strategy that has significantly expanded options for cancer patients. Continuous innovations are improving its precision and effectiveness, making it a vital approach in personalized cancer therapy(8).
To enhance the effectiveness of CAR T-cell therapy and develop better-targeted treatments, researchers are turning to computational techniques like molecular docking. Molecular docking is a method for predicting interactions between a small molecule and a protein at the atomic level with the aid of computers(9). Molecular docking simulations are essential for determining how a drug interacts with its target, ultimately aiding in drug design and optimization(9). This process involves scoring functions to identify the most optimal docked pose, which provides crucial insights into the molecular interactions between the drug and its target. The binding pose is crucial as it reveals how a drug interacts with its target, optimizing binding affinity, specificity, and stability. This knowledge guides drug modifications to enhance efficacy, reduce side effects, and improve overall therapeutic potential. Understanding these interactions allows for the rational design of more effective drugs by optimizing binding affinity, specificity, and stability (9). Molecular docking allows researchers to study how atoms and molecules change and interact over time(10) (9). Molecular dynamics (MD) is a computational technique that predicts the movement of atoms and molecules over time. This information can help us understand the behavior of biological molecules, showing how they change in different conditions and helping understand materials at the atomic level. MD simulations further confirm that the receptors remain intact with each other.
IL-13Rα2 (Interleukin-13 receptor alpha 2) is a key regulator in glioma progression. Researchers are increasingly targeting IL-13Rα2 through therapeutic interventions and gene therapy to improve treatment outcomes. Advanced strategies, including IL-13Rα2-specific CAR-T cells and antibody-based therapies, are being refined to enhance efficacy while preserving brain function (11), Figure 1.
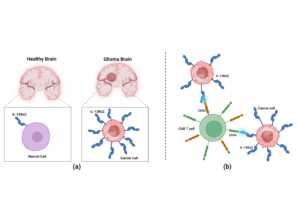
CD45, also known as Leukocyte Common Antigen, is a protein tyrosine phosphatase expressed on all white blood cells. It plays a vital role in immune cell activation, proliferation, and differentiation. Due to its regulatory function in T- and B-cell signaling, CD45 is widely studied in immunology and serves as a critical biomarker in clinical diagnostics(12). CD62 is known as a selectin, a type of cell adhesion molecule responsible for transporting white blood cells during immune responses and inflammation (13). Selectins are of three primary types: E-selectin (CD62E), P-selectin (CD62P), and L-selectin (CD62L)(14). They are each responsible for facilitating leukocyte adhesion, helping leukocyte attachment or rolling, and aiding in migration to lymphoid tissues or inflammatory areas(14).
CD45 and CD62 play essential roles in immune cell signaling and adhesion, making them key targets for immunotherapeutic strategies against glioblastoma. Given their involvement in immune regulation, we hypothesize that the specificity of CAR T-cells targeting IL-13Rα2 may be due to their interactions with CD45 and CD62 receptors. To investigate this, we performed computational simulations to analyze the interactions between CD45, CD62, and IL-13Rα2 on the surface of glioblastoma cells. Our results indicate that CD45-IL-13Rα2 and CD62-IL-13Rα2 strongly interact at the binding site predicted by the graph neural network.
We hypothesize that the particular nature of CAR T-cell could be due to its binding to CD45 and CD62 receptors. Therefore, we have performed computational simulations to study the binding of CD45 and CD62 with IL-13Rα2 located on the surface of glioblastoma cells. Our results show that CD45-IL-13Rα2 andCD62-IL-13Rα2 strongly interact at the binding site predicted by the graph neural network.
MATERIALS AND METHODS.
Receptor structure prediction.
AlphaFold 3 was used to predict CD45, CD62, and IL-13α2 structures, which were imported from the UniProt database (16) (17).
Binding site prediction.
Using the GrASP web server, the possible druggable sites on the surface of IL-13α2, CD45, and CD62 receptors were predicted(18). The HDOCK2.0 server was utilized to simulate docking interactions between CD45-IL-13α2 and CD62-IL-13α2(19). We used an internal script to gain insights into the molecular connections between IL-13α2, CD45, and CD62. This script identified the hydrogen bonds IL-13α2 receptor formed with CD45 and CD62 receptors. This script analyzes hydrogen bonds between two chains in a PDB file using Biopython’s PDB module. It calculates the Euclidean distance between nitrogen (N) and oxygen (O) atoms in different chains and identifies hydrogen bonds if they are within a 4.0 Å cutoff. The results, including residue names, atom names, and bond distances, are printed. The script processes all .pdb files in the current directory.
Molecular docking simulations.
The receptor structures predicted by AlphaFold 3 were used as inputs for the docking process performed by using the HDOCK webserver. The top-ranked docked confirmations based on the docking score were analyzed further.
MD simulations.
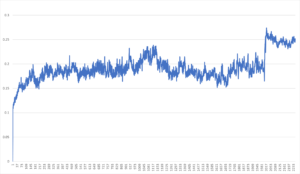
The GROMACS software was utilized to conduct MD simulations to evaluate the dynamics and stability of the CD45-IL-13α2 and CD62-IL-13α2 complexes within a physiological setting(20). Energy minimization and NPT (constant number of particles, pressure, and temperature) equilibration using Berendsen Thermostat (temperature range from 100K-300K) were applied to the complexes derived by HDOCK. After that, each complex underwent 50 ns of production runs. The trajectories were examined for root-mean-square deviation (RMSD) to evaluate the complexes’ stability over time (Figure 2). These simulations’ outcomes provide dynamic insights into the protein complexes’ stability and structural changes. To accurately test the dynamics of CD45-IL-13α2 and CD62-IL-13α2 complexes within a physiological setting, we have used the GROMACS software. The test utilized HDOCK to apply energy minimization and NPT (constant number of particles, pressure, and temperature) equilibration. Afterward, each complex underwent 50 ns of production runs. The binding energy was computed using the PRODIGY web server. The figures were generated using the ChimeraX software.
RESULTS
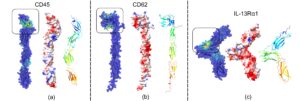
The current research uses computational simulation techniques to understand the interactions between CAR T-cells (containing CD45 and CD62 receptors) and glioma cancer cells (containing IL-13Rα2 receptor). To understand the binding interactions between IL-13Rα2, CD45, and CD62, the “binding sites” were determined (Figure 3). This was done by using a tool known as the GrASP web servers. GrASP web server is a graph neural network-based method that uses artificial intelligence to evaluate electrostatic properties, giving us potential binding sites. The “binding sites”—the most likely locations for binding on these receptors—were initially determined to investigate this interaction. More information, such as the receptors’ electrostatic surface potential (ESP), was also investigated. ESP tells the charge of the protein, in which red represents negative, blue represents positive, and white represents neutral. CD45 was mainly neural based on the ESP, while CD62 and IL-13Rα2 were negatively charged.
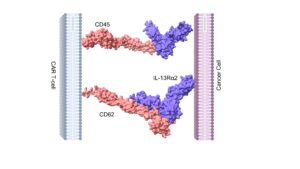
In the following steps, we also computed the binding interactions between the CD45-IL and CD62-IL receptors using molecular docking simulations. Molecular docking is a computational technique that calculates interactions between a small molecule and a larger target. This more profound understanding of the interaction helps future researchers design new drugs and modify existing drugs, making them far more effective. Docking between CD45, CD62, and IL-13Rα2 resulted in two unique complexes, as shown in Figure 4. Results showed that the CD62-IL-13Rα2 complex is partially stronger than the CD45-IL-13Rα2 complex, Table 1. The number of hydrogen bonds showed that CD45 formed a higher number of hydrogen bonds with IL-13α2 in comparison to CD62. Finally, MD simulations were used to further confirm that the two receptors remained bound throughout the entire 50-nanosecond MD simulation. It was observed that both receptors remained bound, further confirming that the receptors interact with each other.
| Table 1. Binding Energy between receptors. | ||
| Binding affinity (kcal/mol) | Number of hydrogen bonds | |
| CD45-IL-13Rα2 | -14.7 | 50 |
| CD62-IL-13Rα2 | -15.4 | 45 |
DISCUSSION.
We performed molecular docking and MD simulations to analyze the binding interactions of CD45 and CD62 receptors on CAR T-cells with the IL-13Ra2 receptor. In addition, we have performed molecular docking and MD simulations to access the receptor interactions.
Based on our results, we found that both receptors, CD45 and CD62, bind strongly to the IL-13Rα2 receptor. Although the binding energy of the CD62-IL-13Rα2 receptor complex was slightly stronger than that of the CD45-IL-13Rα2 receptor complex, this suggests that CD62 may be a more effective binding partner. The binding results were further validated by predicting the protein’s binding site. The predicted binding site was located at the apical region of the receptor, indicating that head-on interactions between the receptors could be possible. To strengthen the findings of this research, experimental validation through in-person laboratory testing would provide additional confirmation of accuracy. While computational methods offer valuable predictive insights, integrating lab-based approaches ensures even greater reliability, particularly when studying cancerous cells. Moreover, to enhance precision, docking simulations can be refined by cross-validating results with multiple docking software. Additionally, binding interaction analyses can complement experimental techniques such as Surface Plasmon Resonance (SPR) and Isothermal Titration Calorimetry (ITC), further enriching the study’s robustness and applicability.
CONCLUSION.
We investigated the interactions between CD45 and CD62 receptors on CAR T-cells and IL-13Rα2 in glioma cells. Our results show that both CD45 and CD62 strongly bind to IL-13Rα2, with CD62 exhibiting slightly higher binding strength and stability. These findings highlight the potential for CD62-based CAR T-cell modifications to enhance glioblastoma targeting. In vitro and in vivo testing are needed to confirm and improve the accuracy of the computational findings. Further advancements in CAR T-cell therapy are required to reduce side effects, increase accuracy, and enhance targeting specificity.
REFERENCES.
- A. Giese, M. Westphal, Glioma invasion in the central nervous system. Neurosurgery. 39, 235-252 (1996).
- A. Shergalis, A. Bankhead, U. Luesakul, N. Muangsin, N. Neamati, Current challenges and opportunities in treating glioblastoma. Pharmacol Rev. 70, 412-445 (2018).
- Y. Yagawa, K. Tanigawa, Y. Kobayashi, M. Yamamoto, Cancer immunity and therapy using hyperthermia with immunotherapy, radiotherapy, chemotherapy, and surgery. Journal of Cancer Metastasis and Treatment. 3, 218 (2017).
- I. Y. Eyüpoglu, M. Buchfelder, N. Savaskan, Surgical resection of malignant gliomas—role in optimizing patient outcome. Nat Rev Neurol. 9, 141-151 (2013).
- P. Dimitrios, Advancements in the Management of Meningioma: A Comprehensive Review of Current Treatment Modalities. Clinics of Oncology. 7, 1-8 (2024).
- M. Koutsioumpa, E. Papadimitriou, Cell surface nucleolin as a target for anti-cancer therapies. Recent Pat Anticancer Drug Discov. 9, 137-152 (2014).
- R. Mohanty et al., CAR T cell therapy: A new era for cancer treatment. Oncol Rep. 42, 2183-2195 (2019).
- E. Rosenbaum et al., Cancer supportive care, improving the quality of life for cancer patients. A program evaluation report. Support Care Cancer. 12, 293-301 (2004).
- J. Fan, A. Fu, L. Zhang, Progress in molecular docking. Quantitative Biology. 7, 83-89 (2019).
- J. B. Ghasemi, A. Abdolmaleki, F. Shiri, in Pharmaceutical sciences: breakthroughs in research and practice. (IGI Global, 2017), pp. 770-794.
- C. Xu et al., IL-13Rα2 humanized scFv-based CAR-T cells exhibit therapeutic activity against glioblastoma. Mol Ther Oncolytics. 24, 443-451 (2022).
- M. L. Hermiston, Z. Xu, A. Weiss, CD45: a critical regulator of signaling thresholds in immune cells. Annu Rev Immunol. 21, 107-137 (2003).
- A. Aruffo, W. Kolanus, G. Walz, P. Fredman, B. Seed, CD62/P-selectin recognition of myeloid and tumor cell sulfatides. Cell. 67, 35-44 (1991).
- G. S. Gupta. L-selectin (CD62L) and its ligands. Immunity. 14, 553-574 (2012).
- C. E. Brown et al., Locoregional delivery of IL-13Rα2-targeting CAR-T cells in recurrent high-grade glioma: a phase 1 trial. Nature Medicine. 30, 1001-1012 (2024).
- J. Abramson et al., Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature. 1-3 (2024).
- E. Boutet, D. Lieberherr, M. Tognolli, M. Schneider, A. Bairoch, in Plant bioinformatics: methods and protocols. (Springer, 2007), pp. 89-112.
- Z. Smith, M. Strobel, B. P. Vani, P. Tiwary, Modeling, Graph attention site prediction (grasp): Identifying druggable binding sites using graph neural networks with attention. J Chem Inf Model. 64, 2637-2644 (2024).
- Y. Yan, H. Tao, J. He, S.-Y. Huang, The HDOCK server for integrated protein–protein docking. Nat Protoc. 15, 1829-1852 (2020).
- D. Van Der Spoel et al., GROMACS: fast, flexible, and free. Journal of computational chemistry 26, 1701-1718 (2005).
Posted by buchanle on Thursday, June 19, 2025 in May 2025.
Tags: CAR T-cell Therapy, Glioma, Molecular Docking, Molecular Dynamics

