Application of Machine Learning and Deep Learning in Brain tumor detection in MRI scans – Comparison and Analysis of different models
ABSTRACT
This study explores the application of machine learning and deep learning models to detect and classify brain tumors in MRI scans, aiming to enhance diagnostic accuracy and efficiency. Recognizing the challenges radiologists face—such as time-consuming analysis and the risk of missed tumors—we employed two distinct datasets: one for binary classification (tumor presence) with 253 low-resolution images and another for multiclass classification (tumor type) with 5,712 high-resolution images of pituitary tumors, gliomas, meningiomas, and controls. We trained and evaluated multiple machine learning algorithms—including Random Forest, K-Nearest Neighbors (KNN), Logistic Regression, and Gaussian Naive-Bayes—as well as neural networks like Multi-Layer Perceptron (MLP) and Convolutional Neural Networks (CNN). CNNs showed significant potential due to their ability to extract spatial features from images, achieving the highest accuracy of 90.2% for binary and 88.9% for multiclass classification. The Random Forest classifier also achieved high accuracy and strong recall rates, indicating robust tumor detection capabilities. KNN exhibited the highest sensitivity, thus minimizing false negatives. These results suggest that integrating machine learning models like Random Forest and CNNs into clinical practice could greatly help radiologists accurately detect and classify brain tumors, resulting in improved patient outcomes.
INTRODUCTION.
A brain tumor is an abnormal growth of cells in or around the brain. It can be benign (non-cancerous) or malignant (cancerous). Regardless of its benign or malignant nature, a brain tumor may alter brain function, thereby impacting health. It may present itself through headaches or seizures and can lead to paralysis or even death. The survival rate of patients diagnosed with brain tumors depends on the tumor’s location and grade. The mean five-year survival rate in the US population with malignant tumors is 33.4% [1].
Radiologists worldwide primarily use images from Magnetic Resonance Imaging (MRI) scans to detect tumors in the brain. However, detecting brain tumors is a complex and time-consuming task. Radiologists are often burdened with long queues of scans to read. This results in them having to go through many normal (non-tumor) scans before coming across a case that has a tumor. Ideally, radiologists would prefer to quickly dispose of negative cases and then spend more time on positive cases to make a specific diagnosis and classify the tumor.
Artificial intelligence (AI) can be a vital tool for radiologists to overcome the above challenges. It can quickly identify supposed positive cases and ‘flag’ them so that radiologists can prioritize their reading of these cases, spending more time making a diagnosis and grading the tumor.
Currently, most research on utilizing Artificial Intelligence to identify brain tumors focuses exclusively on neural networks [2]. However, this approach is inadequate to truly identify the most suitable model for clinical use. For this, an evaluation of the performance of these neural networks is needed, along with their comparison to conventional Machine Learning models. Additionally, there is a need to test these models on a large dataset, similar to a real-life clinical setting, in order to ensure their performance scales with larger amounts of data.
The above two objectives are the focus of our research. We used two datasets, one small and one large, to compare and analyze the various models and determine the most effective overall method for brain tumor detection and classification, employing metrics such as overall accuracy [3] and recall score [4].
MATERIALS AND METHODS.
Implementation.
All models in this study were trained and tested on Google Colab, an online hosted Jupyter Notebook service that provides access to computing resources, including CPUs and GPUs. Specifically, a runtime on the Google Colab website was created using Python 3.10 with the following resources: T4 GPU, 13 GB System RAM, and 112 GB disk.
Datasets.
This research was carried out in two steps, using two distinct datasets. The first dataset, which was used for binary classification (screening for positive vs negative cases) of axial MRI images, was provided by Inspirit AI Inc. It contained 253 axial images. The images in this dataset were of two types – 155 positive images (containing a brain tumor) and 98 negative images (without any brain tumor). 80% of these images (202 images) were used to train the models, while the remaining 20% (51 images) were set aside for testing. This dataset consisted of low-resolution images for a faster runtime. The second dataset [5] was larger. It was used to broadly classify brain tumors into three types – Pituitary tumor, Intra-axial brain tumor (Glioma), or Meningioma, and was a combination of publicly available health datasets taken from the online Machine Learning library – Kaggle [6]. It consisted of 5,712 images of four types – 1,457 images containing a pituitary tumor, 1321 images of intra-axial brain tumors (Gliomas), 1,339 images of Meningioma, and 1,595 images containing no tumor (control). 80% of these images (4,570 images) were used to train the models, while the remaining 20% (1,142 images) were set aside for testing. This dataset consisted of high-resolution images to improve the accuracy of classification. A summary of these datasets is provided in Table 1.
| Table 1. Summary of the datasets used. | |||
| Dataset | No. of Images | Categories | Use |
| Dataset 1 | 253 | Tumor | Binary screening/ Flagging |
| No Tumor | |||
| Dataset 2 | 5712 | Pituitary Tumor | Classification into tumor types |
| Meningioma | |||
| Intra-axial tumor(Glioma) | |||
| No tumor | |||
Methodology.
The first dataset – to be used for binary classification – was preprocessed and split for training (80%) and testing (20%). Multiple ML models and Neural Networks were trained and tested on this dataset. The accuracy of these different models was compared to select the best-performing model/network for the study. Then, a second dataset – to be used for broad classification into three brain tumor types – was preprocessed and split for training (80%) and testing (20%). The same steps were also carried out for the second dataset.
Data Preprocessing.
The images in both datasets had to be optimized before being used in ML models. The first step involved changing the images to grayscale (1 color channel) from Red, Green, and Blue (3 color channels). No data was lost during this step and it was done to decrease the complexity in the design of the ML models, resulting in faster training. Grayscaling was done by accessing the color processing functionalities provided by the color module within the skimage package.
In the second step, the range of pixel intensity values was made constant across all images to be between 0 and 1 (normalization) (Fig. 1). This was done to ensure that the algorithm would not overfit based on unrelated differences between classes, such as different brightnesses, which can be technical artefacts rather than an actual tumor. [7, 8]
Third, data augmentation was performed on the first dataset to double the number of images by transforming (horizontally flipping) the pre-existing ones (Fig. 2). This was done to help the ML models learn to detect tumors irrespective of their location in the brain or the scan. The images in the second dataset were cropped to eliminate the blank background as much as possible by automatically detecting the edges of the brain scan.
In the final preprocessing step, all images were flattened before being used for training and testing. Flattening involved converting the two-dimensional image into a one-dimensional array so the ML models could process it.
Machine Learning Models Used.
Random Forest Classifier. Random Forest is a robust ensemble learning algorithm widely employed in machine learning for classification and regression tasks. Its advantage is that it utilizes a collection of decision trees, each contributing to its prediction. By exploiting the wisdom of multiple trees, Random Forest alleviates the risk of overfitting and enhances predictive accuracy [9]. It was imported from the sklearn.ensemble package.
K-Nearest Neighbors (KNN). The K-Nearest Neighbors (KNN) algorithm is a straightforward ML technique for classification and regression. It stores all available cases and classifies new instances based on a similarity measure (e.g., distance functions). K-NN identifies the ‘k’ closest data points in the training set for new input and assigns the input to the most common category [10]. The classifier was imported from the sklearn.neighbors package.
Logistic Regression. Logistic regression is a statistical method used for binary classification problems where the goal is to predict one of two possible outcomes. It models the probability that a given input belongs to a particular category by fitting a logistic function (sigmoid) to the data. The algorithm estimates the coefficients for each feature through maximum likelihood estimation, transforming the input features into a value between 0 and 1. This probability is then thresholded to decide the class label: values above 0.5 indicate one class, while values below indicate the other [11]. It is imported from the sklearn.linear_model package.
Gaussian Naive-Bayes (GNB). Gaussian Naive Bayes assumes a bell-shaped distribution in the data. It calculates the probability of each class for unseen data using these distributions and Bayes’ theorem. A downside to this algorithm is its assumption of independent features, which may not always hold. GaussianNB was imported from the sklearn.naive_bayes package.
Neural Networks Used.
Multi-Layer Perceptron (MLP). A Multi-Layer Perceptron (MLP) is a computational model inspired by how biological neural networks work in the human brain. It consists of layers of interconnected nodes (neurons), where each connection has an associated weight. The network typically has an input layer, one or more hidden layers, and an output layer. When data is fed into the input layer, it gets processed through the hidden layers, where the neurons apply a weighted sum of their inputs followed by a non-linear activation function [12]. The final output layer produces the prediction. It was imported from the sklearn.neural_network package.
Convolutional Neural Network (CNN). A Convolutional Neural Network (CNN) is a specialized type of Artificial Neural Network primarily used for processing and analyzing visual data. It consists of multiple layers, including convolutional, pooling, and fully connected layers. In convolutional layers, the network uses filters (kernels) to scan the input image and produce feature maps, detecting patterns such as edges and textures. Pooling layers then down sample these feature maps to reduce dimensionality. The final classification is performed by the fully connected layers at the end, using the extracted features from the image [13]. The structure of the best-performing CNN is given in the results section.
Understanding The Output Metrics.
This research was done to have maximum sensitivity. Sensitivity measures the proportion of true positive cases (brain tumors) correctly identified by the model. It is also known as the true positive rate or recall; specificity, however, measures the proportion of true negative cases (healthy cases) correctly identified, indicating how well the model avoids false positives.
In brain tumor detection, missing a tumor (a false negative) could have severe consequences, so the models and neural networks used were compared based on having the best balance between high sensitivity and accuracy: the best model minimizes the risk of missing a tumor.
The ML models’ predictions were tested by calculating the accuracy and recall scores. The accuracy percentage measures the proportion of correct predictions, while the recall percentage measures the proportion of true positives correctly identified by the model.
RESULTS.
The research presented in this study demonstrates the practical application of machine learning (ML) and deep learning models for detecting and classifying brain tumors using MRI images.
ML Models.
The accuracies of the machine learning models after training on the first dataset are given in Table 2. Random Forest Classifier achieved the highest overall accuracy of 89.5% on the first dataset, while K-Nearest Neighbors achieved the highest recall of 91%.
| Table 2. Accuracies of the ML Models on the first dataset. | ||
| Model Name | Accuracy (%) | Recall (%) |
| Random Forest | 89.5 | 87.5 |
| K-Nearest Neighbors (KNN) | 86.2 | 91 |
| Logistic Regression | 82.3 | 87 |
| Gaussian Naïve-Bayes (GNB) | 71.5 | 73.2 |
The accuracies of the machine learning models after training on the second dataset are given in Table 3. Random Forest again achieved the highest accuracy (86.9%) and also the highest recall of 87.5%.
| Table 3. Accuracies of the ML Models on the second dataset. | ||
| Model Name | Accuracy (%) | Recall (%) |
| Random Forest | 86.9 | 87.5 |
| K-Nearest Neighbors (KNN) | 82.9 | 82.9 |
| Logistic Regression | 79.2 | 79 |
| Gaussian Naïve-Bayes (GNB) | 65.8 | 68.1 |
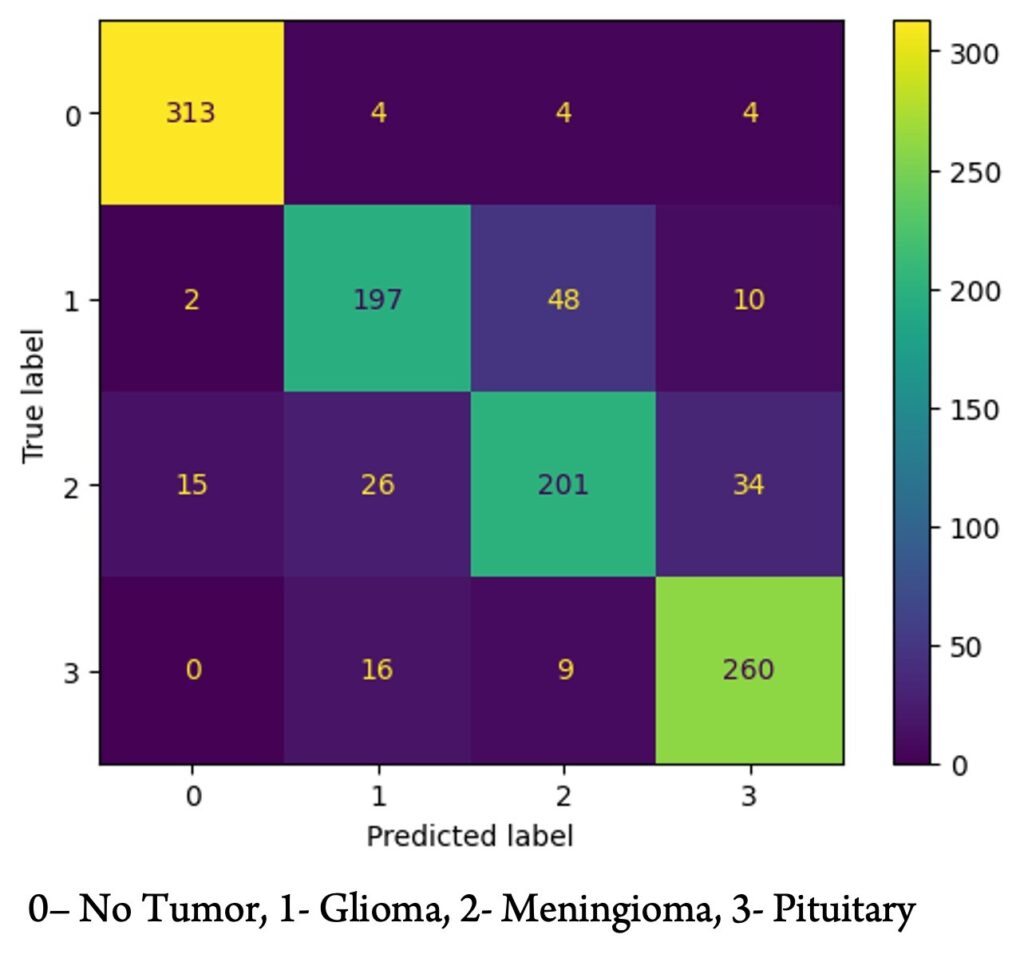
Figure 3 shows the confusion matrix of the Random Forest model after training on the second dataset. It is evident that the model was proficient in distinguishing whether a scan contained a tumor but had more difficulty differentiating between gliomas and meningiomas.
Neural Networks.
The Convolutional Neural Network (CNN) performed the best out of all the models tested on both datasets. A sample structure of the best-performing CNN on the first dataset is given in Table 4.
| Table 4. Structure of the best-performing CNN on Dataset 1 | ||
| Layer (type) | Output Shape | Parameters |
| conv2d (Conv2D) | (None, 75, 75, 64) | 320 |
| max_pooling2d | (None, 37, 37, 64) | 0 |
| flatten_1 (Flatten) | (None, 87616) | 0 |
| dense (Dense) | (None, 64) | 5607488 |
| dropout_1 (Dropout) | (None, 64) | 0 |
| dense_1 (Dense) | (None, 128) | 8320 |
| dropout_2 (Dropout) | (None, 128) | 0 |
| dense_5 (Dense) | (None, 2) | 258 |
This model had eight layers, with one convolutional and three dense layers. The total numbers of parameters were 5616386, totalling to 21.42 MB. It achieved a maximum accuracy of 90.2%.
Similarly, on the second dataset, a CNN (with a different structure) achieved a maximum accuracy of 88.9%.
Multi-Layer Perceptron had a lower overall accuracy of 80%.
DISCUSSION.
Performance of the ML Models.
The performance of different machine learning algorithms varied across both datasets. For the binary classification task (Dataset 1), the Random Forest classifier achieved the highest overall accuracy of 89.5% and a recall of 87.5%. This suggests that Random Forest was adept at identifying the presence of brain tumors, making it a strong candidate for screening purposes. KNN, on the other hand, exhibited the highest recall score (91%), indicating that it was the most sensitive model in detecting tumors. Sensitivity is particularly critical in this context because missing a tumor (a false negative) can have severe consequences for patient outcomes. Logistic regression also performed well, though it fell short of the accuracy and recall metrics of Random Forest and KNN.
For the second dataset, which focused on classifying tumor types, Random Forest once again emerged as the top-performing model with an accuracy of 86.9% and a recall of 87.5%. KNN was second with 82.9% accuracy and recall, while other models like Logistic Regression and Gaussian Naive-Bayes had inferior results. These results emphasize the capability of Random Forests in handling complex classification tasks, such as differentiating between different types of brain tumors.
The Gaussian Naive-Bayes model performed the worst across both datasets, with accuracy rates of 71.5% and 65.8% for binary and tumor type classification, respectively. Given the complex relationships between different features in MRI images, the assumption of independent features in Naive-Bayes likely contributed to its suboptimal performance.
Performance of the Neural Networks.
The study’s implementation of neural networks, specifically the Multi-Layer Perceptron (MLP) and Convolutional Neural Network (CNN), yielded varying results. The MLP, with a single hidden layer of size 200, could not achieve a high accuracy because it only had a single hidden layer. However, the CNN demonstrated the most potential due to its ability to extract and analyze spatial features from MRI images. It performed the best overall on both datasets, with an accuracy of 90.2% for binary-type classification and 88.9% for tumor-type classification. Also, the best-performing CNN architecture, consisting of multiple layers, such as convolutional, pooling, and dropout layers, achieved high accuracy with a relatively small size (~21.42 MB). This efficiency in both processing power and accuracy makes CNNs suitable for future brain tumor detection and classification developments. Similar findings were observed by Assel Ibraimova et al. [14] and Peiyi Gao et al. [15], who investigated the potential of using convolutional neural networks (CNNs) for diagnosing brain diseases based on MRI scans. However, they did not compare CNN with conventional machine learning methods and used data from a single center.
Implications for Clinical Use.
The findings of this research are significant for the use of AI in clinical radiology. By using machine learning algorithms such as Random Forest and deep learning techniques like CNNs, radiologists can significantly improve the speed and accuracy of brain tumor detection. Particularly in high-throughput settings where radiologists must review numerous scans, AI tools could help prioritize cases with potential tumors, allowing radiologists to focus their attention on the most critical scans. Zhihua Liu et al. [16] have provided a comprehensive survey of deep learning-based brain tumor segmentation techniques with similar findings to our research.
Another critical benefit of AI is the potential to reduce false negatives (missed tumors). Models such as KNN, which demonstrated high recall, can be beneficial in flagging even small or atypical tumors that radiologists might otherwise miss in routine settings. Also, by integrating tumor classification algorithms, AI systems could suggest whether a tumor is likely a pituitary tumor, glioma, or Meningioma.
Limitations.
Despite the promising results, several limitations exist in this study. First, Dataset 1 was relatively small, containing only 253 images. While the models performed well in this limited context, their performance might only generalize to larger, more diverse datasets like Dataset 2, with further training and validation.
Second, while the models focused on sensitivity, an additional focus on specificity would be beneficial to reduce the number of false positives. In a clinical context, false positives can lead to unnecessary follow-up procedures, increased patient anxiety, and higher healthcare costs.
Future Work.
In future research, a larger dataset with images from different sources and varied patient demographics could be used to enhance the models’ accuracy and relevance to real-world scenarios. Future studies could improve the balance between sensitivity and specificity, ensuring that AI models not only detect tumors, but also accurately differentiate between benign and malignant lesions.
CONCLUSION.
In conclusion, this study demonstrates the significant potential of machine learning and deep learning algorithms in improving brain tumor detection and classification from MRI scans. By training various models on two datasets—one for binary classification (tumor vs. no tumor) and another for multiclass classification of gliomas, meningiomas, and pituitary tumors—we found that Convolutional Neural Networks (CNNs) achieved the highest accuracies (90.2% and 88.9%, respectively), demonstrating their potential in extracting spatial features. Among simple ML models, Random Forest had the highest accuracy, while K-Nearest Neighbors (KNN) showed the highest sensitivity at 91%, which is beneficial for decreasing false negatives. These results show that combining models like CNNs and Random Forest into radiological practice can enhance diagnostic accuracy and efficiency in radiology. Future research with larger datasets could further validate these models for real-world applications.
ACKNOWLEDGMENTS.
I want to express my gratitude to Ms. Shreya Parchure, MD-PhD student at the University of Pennsylvania, for her unwavering support, guidance, and resources throughout my research journey.
REFERENCES
- National Cancer Institute. “Cancer Stat Facts: Brain and Other Nervous System Cancer.” Accessed January 22, 2024. https://seer.cancer.gov/statfacts/html/brain.html.
- M. Sharma, N. Miglani. Automated brain tumor segmentation in MRI images using deep learning: overview, challenges and future. Deep learning techniques for biomedical and health informatics, 347-383 (2020).
- Scikit-learn. “accuracy_score.” Accessed June 29, 2024. https://scikit-learn/stable/modules/generated/sklearn.metrics.accuracy_score.html.
- Scikit-learn. “recall_score.” Accessed June 29, 2024. https://scikit-learn/stable/modules/generated/sklearn.metrics.recall_score.html.
- Kaggle. “Brain Tumor MRI Dataset.” Accessed February 3, 2024. https://www.kaggle.com/datasets/masoudnickparvar/brain-tumor-mri-dataset.
- Kaggle. “Find Open Datasets and Machine Learning Projects.” Accessed February 3, 2024. https://www.kaggle.com/datasets.
- M. Kociołek, M. Strzelecki, R. Obuchowicz, Does image normalization and intensity resolution impact texture classification. Computerized Medical Imaging and Graphics 81, 101716 (2020).
- A. Carré, G. Klausner, M. Edjlali, and others, Standardization of brain MR images across machines and protocols: bridging the gap for MRI-based radiomics. Scientific Reports 10, 12340 (2020).
- L. Breiman, Random forests. Machine Learning 45, 5–32 (2001).
- Z. Zhang, Introduction to machine learning: k-nearest neighbors. Annals of Translational Medicine 4, 218 (2016).
- D. W. Hosmer Jr, S. Lemeshow, R. X. Sturdivant, Applied Logistic Regression (John Wiley & Sons, 2013).
- Bengio, Y., Ducharme, R., Vincent, P. A neural probabilistic language model. Adv. Neural Inf. Process. Syst. 13, (2000).
- K. Fukushima, Neocognitron: A self-organizing neural network model for a mechanism of pattern recognition unaffected by shift in position. Biol. Cybernetics36, 193–202 (1980).
- Ibraimova, A., Satybayeva, A., Neverova, Y., Sarsenbayev, B., Toxanova, S. Automated screening of brain disorders: A machine learning model for MRI classification. Int. J. Commun. Netw. Inf. Secur. 15, 118–133 (2023).
- P. Gao, W. Shan, Y. Guo, and others, Development and validation of a deep learning model for brain tumor diagnosis and classification using magnetic resonance imaging. JAMA Network Open 5, e2225608 (2022).
- Liu, Z., Tong, L., Chen, L., Jiang, Z., Zhou, F., Zhang, Q., Zhang, X., Jin, Y., Zhou, H. Deep learning-based brain tumor segmentation: A survey. Complex Intell. Syst. 9, (2022).
Posted by buchanle on Thursday, June 19, 2025 in May 2025.
Tags: Brain tumors, Deep Learning (DL), Machine Learning (ML), Neural Networks

