Analyzing the role of endothelial cell derived Jedi-1 in postnatal neurogenesis
ABSTRACT
Impaired neurogenesis has been implicated in the development of many neurodegenerative diseases. The proliferation of neural stem cells (NSCs) must be efficiently controlled to be able to generate new neurons in the event of these diseases, brain injury, and to support development. Jedi-1, a newly discovered protein present in the neurogenic subventricular zone in mice, has been shown to suppress the proliferation of NSCs. However, the mechanism of this regulation is not known. The effects of Jedi-1 are non-cell autonomous, since it is not expressed by NSCs. This study seeks to better characterize the role of Jedi-1 in neurogenesis. Immunostaining revealed that Jedi-1 is expressed in blood vessels in the neurogenic niche and co-culturing brain endothelial cells with NSCs demonstrated that these vascular cells significantly reduce the proliferation rate of NSCs. These results suggest that Jedi-1 regulates neurogenesis through its action in local vascular endothelial cells. Future studies are will directly determine whether loss of Jedi-1 in endothelial cells affects NSC proliferation.
INTRODUCTION.
The proliferation of neural stem cells (NSCs) allows new neurons to be generated for development, injury recovery, and many other important processes [1]. This proliferation must be efficiently regulated in order to ensure appropriate function of the brain in every stage of life. Importantly, impaired neurogenesis has been implicated in a multitude of neurodegenerative diseases [2]. Neurogenesis occurs in two distinct areas in the brain: the ventricular-subventricular zone (V-SVZ) of the lateral ventricles and the subgranular zone (SGZ) of the hippocampus [3]. These neurogenic regions consist of four main cell subtypes: quiescent NSCs, active NSCs, progenitor/intermediate cells, and neurons [1]. The vital process of neurogenesis is modulated by an array of factors that work together to create an extensive microenvironment to support NSCs; however, a majority of these regulating factors are poorly understood.
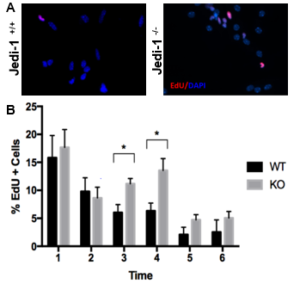
Preliminary data from our laboratory suggests that Jedi-1 (also known as PEAR1), a surface protein present in various cell types in the nervous system, plays a role in the regulation of neurogenesis in the rodent brain. This protein was first identified in mice as a phagocytic receptor present in glial cells in the dorsal root ganglia, and it functions primarily to recognize apoptotic neurons during development that need to be disposed to avoid inflammation [4]. The role of Jedi-1 in the central nervous system has not been characterized, especially in regards to its effect on neurogenesis. Previous studies have shown that phagocytosis of apoptotic cells in the stem cell niche by precursor cells is necessary to maintain the proliferation of neural stem cells [3]. These studies led to preliminary research on the possible relationship between Jedi-1 and regulation of neurogenesis, which showed that the loss of Jedi-1 increases proliferation of NSCs (Figure 1). Interestingly, unlike other regulatory factors, Jedi-1 protein was not detected in the NSCs themselves but was expressed in endothelial cells (ECs) of blood vessels adjacent to the V-SVZ (Figure S1). This observation of increased proliferation in the absence of endothelial Jedi-1 is in agreement with other studies that have shown that factors released by endothelial cells can have an inhibitory effect on neurogenesis [6, 7].
To better understand the mechanism by which neurogenesis is regulated, this research investigated the specific role of Jedi-1 in NSC proliferation dynamics, helping clarify the role of endothelial Jedi-1 in NSC regulation. Previous research indicating the presence of Jedi-1 in endothelial cells combined with the ability of ECs to promote quiescence alludes to the possibility of endothelial cell Jedi-1 having the ability to regulate neurogenesis (Figure 1). To test this hypothesis, this study quantified the incorporation of EdU, a cell proliferation marker, in NSCs cultured with a Jedi-1 positive brain endothelial cell line and compared it to NSC proliferation in the presence of mouse embryonic fibroblasts (as a control). Further understanding of the signaling pathways that regulate neurogenesis will help direct research in the direction of discovering better regenerative treatments for neurodegenerative disorders.
Figure 1. NSCs isolated from WT mice exhibit higher proliferation compared to Jedi-1 knockout mice. Representative microscopy images (A) of the WT and KO cultures are shown. The bar graph (B) represents the quantification of EdU uptake by cells per field (mean ± SEM.). (Carter lab, unpublished data).
MATERIALS AND METHODS.
bEnd.3 Culturing.
bEnd.3, a murine brain endothelial cell line obtained from the American Type Culture Collection (ATCC), was cultured in T-25 flasks with Dulbecco’s Modified Eagle Medium (DMEM) media supplemented with 10% Fetal Bovine Serum (FBS). Cells were split regularly using Trypsin as the dissociation reagent to avoid high confluence, and all experiments were conducted before the tenth passage in order to minimize senescent cells. Cells were counted using a hemocytometer and plated onto collagen-coated, acid-washed coverslips as required for each experiment. One passage prior to use in the co-culture experiment, the cells were habituated in differentiation media from the NeuroCultTM differentiation kit. Identical procedures were used for Mouse Embryonic Fibroblasts (MEFs), the negative control used for the co-culture experiment.
Jedi-1 Immunocytochemistry Assay.
bEnd.3 cells were plated onto coverslips, along with Jedi-1 transfected HeLa cells and untransfected HeLa cells as positive and negative controls. Three coverslips were replicated for each condition. All cell types were plated at a density of 50,000 cells/ coverslip. The cells were fixed in 10% neutral buffered formalin (NBF) for 15 minutes and fixed then stained using standard techniques. Primary antibody against Jedi-1 (R&D AF7607) and the corresponding secondary antibody Alexafluor 568 (abcam AB175712) was used to identify the localization of protein expression. The cells were mounted in Prolong Gold (ThermoFisher P36931) mounting media with DAPI to mark nuclei.
Generating Neurospheres.
Ventricular-subventricular zone tissue was microdissected from coronal brain sections of postnatal day four C57BL/6 mice. Tissue from 4 pups were dissected and cultured for each experiment. Tissue sections posterior to the olfactory bulbs and anterior to the hypothalamus were used. The dissected tissue was dissociated and plated in uncoated 10 cm2 petri dishes in proliferation media using the NeuroCultTM were allowed to proliferate as spheres for 7-10 days before being dissociated again using the NeuroCultTM differentiation kit, and then dyed with CellTracker Orange.
Labeling NSCs with CellTrackerTM Dye.
Immediately prior to the co-culture experiment, dissociated NSCs were washed with NeuroCultTM base media to remove residual serum. Washed NSCs were incubated in a 2 µM CellTracker Orange dye solution in base media for 30 minutes at 37°C. Cells were harvested by centrifugation to form a pellet. The resultant pellet was re-washed in base media then suspended in desired media and plated at required concentrations.
Co-culture.
bEnd.3 cells were plated on collagen coated coverslips in a 24-well plate at a density of 100,000 cells/well. Directly after bEnd.3 plating, NSCs dyed with CellTracker Orange dye were plated on top of the endothelial cells at 50,000 cells/well. Differentiation media was used to complete the 500 μL total well volume. The co-culture was incubated for three days before being pulsed with EdU at 0.5 μL EdU per 500 μL media for 30 minutes, fixed in NBF and labelled with 5-isomer FAM azide. Coverslips were mounted in Prolong Gold mounting media with DAPI. For controls, conditions of NSCs without CellTracker Orange, co-cultures not pulsed with Edu, and cells with neither CellTracker Orange nor EdU were used. These conditions were replicated for 3 coverslips each, and the entire experiment was replicated using MEFs in the place of bEnd.3 cells.
Imaging and Statistical Analysis.
All imaging was done using a Nikon Eclipse Ti inverted epifluorescence microscope, at 20x magnification. Images were compiled and analyzed using ImageJ. Statistical differences between samples were analyzed using a Student t-test at α=0.05 for n=3 samples.
RESULTS.
Brain endothelial cells express Jedi-1.
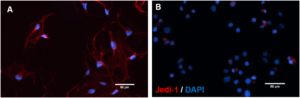
b.End3 cells were cultured to test the role of endothelial Jedi-1 in regulating NSC proliferation. To ensure that the cell line is a good candidate for this experiment, we tested for expression of Jedi-1 protein in the cultured bEnd.3 cells. b.End3 cells were cultured in standard conditions, fixed, and immunolabelled for Jedi-1 (Figure 2A). The process was replicated with HeLa cells as a negative control for nonspecific antibody binding (Figure 2B). In order to confirm our method was successful, Jedi-1 transfected HeLa cells were stained as a positive control (Figure S2).
Figure 2. b.End3 cells express Jedi-1: Representative images of b.End3 cultures immunolabeled with Jedi-1 (red) and DAPI (blue). [A] bEnd.3 [B] HeLa (negative control). Scale bar = 50 μm. n =2.
These cells exhibit robust Jedi-1 expression localized mainly in the membrane and the processes, compared to no expression in the HeLa control experiment (Figure 2).
Presence of Jedi-1+ endothelial cells decreases proliferation of NSCs.
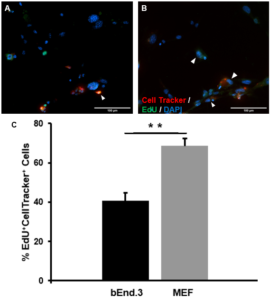
A co-culture experiment was conducted to test the effect of Jedi-1 positive bEnd.3 cells on the proliferation of NSCs. Incubation of differentiating NSC cultures with bEnd.3 cells significantly reduced the proliferation of NSCs. When cultured with MEFs, the baseline amount of proliferating NSCs was observed to be 68.63 ± 6.25% (Figure 3B). When cultured with bEnd.3 cells, there was a significant reduction (p=0.003) in the amount of proliferating NSCs at 40.75 ± 7.64% (Figure 3A, 3C).
Figure 3. Jedi-1+ bEnd.3 cells reduce the number of proliferating NSCs: Representative images of co-cultures stained with DAPI (blue), EdU (green), and CellTracker Orange (Red). [A] bEnd.3 and NSCs co-culture [B] MEF and NSC culture as reference point for proliferation in the absence of Jedi-1. Scale bar = 50 μm. n = 3. Arrowheads indicate EdU+CellTracker cells. [C] The bar graph represents a quantification of the fraction of EdU+CellTracker+ relative to total CellTracker+ cells (mean ± SEM).
DISCUSSION.
Although well known to be integral in many of life’s important processes, the mechanisms of neurogenesis regulation have yet to be fully characterized. These findings suggest that Jedi-1, a surface receptor protein identified in mouse models, may be a previously unrecognized factor contributing to this regulation. This research confirms that endothelial cells, which form vascular structures in the brain, express Jedi-1, a conclusion which is in agreement with other studies that have identified Jedi-1 expression in vascular structures in other areas of the body [5]. Furthermore, this study found preliminary results which suggest that NSC proliferation is decreased in the presence of Jedi-1+ endothelial cells. This result of decreased neurogenesis due to endothelial factors is supported by other studies, namely, one that suggests that endothelial NT-3 promotes quiescence in the V-SVZ [6]. Combined, these two experiments give supporting evidence to the hypothesis that endothelial-derived Jedi-1 helps to regulate neurogenesis by promoting a quiescent NSC state.
Currently, the expression of EdU in co-culture do not show localization around the nuclei, a characteristic common to traditional proliferation assays [3, 6]. This could possibly be attributed to the inclusion of the FAM azide in the EdU reaction buffer as it had previously never been used by my lab before. Preliminary results using the FAM azide appear to show faint expression compared to other azides. Future optimization of this assay is necessary to confirm the results of this experiment. The role of Jedi-1 in neurogenesis dynamics will be further characterized in vitro through Jedi-1 knockdown in bEnd.3 cells and by using primary Jedi-1-/- vs +/+ endothelial cells. In doing this, non-Jedi-1 related effects on neurogenesis will be accounted for, allowing specific endothelial-derived Jedi-1 results to be analyzed. A similar endothelial Jedi-1 conditional knock out will be conducted in vivo to confirm these results. Furthermore, the co-culture experiment will be done to examine the effect of endothelial-derived Jedi-1 on KO NSCs compared to WT NSCs used in this experiment. These results will allow the effect of Jedi-1 to be analyzed when there are pre-existing varied rates of proliferation to understand if Jedi-1 has a more significant impact when proliferation is occurring at higher rates. Finally, it will be important to determine what factors are released by the endothelial cells in the presence and absence of Jedi-1 and identify those affecting neurogenesis.
By identifying this role of EC-derived Jedi-1, the overall process of neurogenesis regulation can be better understood. More efficient neurogenesis would allow for better treatment against signs of neurodegenerative disease and aging, processes in which neurogenesis plays an important role [1, 2]. By promoting the quiescence of NSCs, the NSC pool can be better maintained to ensure that NSCs will be present throughout the duration of one’s life in the event of an injury or other situation which required the generation of new neurons. Ultimately, this research contributes to a more widespread effort to better understand how the brain functions and how to keep those functions as efficient as possible.
ACKNOWLEDGMENTS.
I would like to thank Dr. Bruce Carter, Ketaki Katdare, Vivianne Morrison, Alex Trevisan, and everyone at the Carter lab for giving me the opportunity to conduct research and helping me throughout the entire process of learning and experimenting. I would also like to thank Dr. Deweese and the School for Science and Math at Vanderbilt for being continuous advisors throughout my experience in the lab. Lastly, I would like to acknowledge the National Institutes of Health for funding this research (R01 NS102365).
SUPPORTING INFORMATION.
Supplemental Figure S1
REFERENCES.
- L. Katsimpardi, P. M. Lledo, Regulation of neurogenesis in the adult and aging brain. Current Opinion in Neurobiology. 53, 131-138 (2018).
- B. Winner, Z. Kohl, F. H. Gage, Neurodegenerative disease and adult neurogenesis. European Journal of Neuroscience. 33, 1139-1151 (2011).
- Z. Lu, M. R. Elliot, Y. Chen, J. T. Walsh, A. L. Klibanov, K. S. Ravichandran, J. Kipnis, Phagocytic activity of neural progenitors regulates adult neurogenesis. Nature Cell Biology. 13, 1076-1083 (2011).
- H. Wu, E. Bellmunt, J.L. Scheib, V. Venegas, C. Burkert, L. F. Reichardt, Z. Zhou, I. Fariñas, B. D. Carter, Glial precursors clear sensory neuron corpses during development via Jedi-1, an engulfment receptor. Nature Neuroscience, 12, 1534-1541 (2009).
- N. Nanda, M. Bao, H. Lin, K. Clauser, L. Komuves, T. Quertermous, P. B. Conley, D. R. Phillips, M. J. Hart, Platelet Endothelial Aggregation Receptor 1 (PEAR1), a Novel Epidermal Growth Factor Repeat-containing Transmembrane Receptor, Participates in Platelet Contact-induced Activation. Journal of Biological Chemistry, 12, 24680-24689 (2005).
- A. C. Delgado, S. R. Ferrón, D. Vicente, E. Porlan, A. Perez-Villalba, C. M. Trujillo, P. D’Ocón, I. Fariñas, Endothelial NT-3 Delivered by Vasculature and CSF Promotes Quiescence of Subependymal Neural Stem Cells through Nitric Oxide Induction. Neuron, 83, 572-585 (2014).
- T. Licht, E. Keshet, The vascular niche in adult neurogenesis. Mechanisms of Development, 138, 56-62 (2015).
Posted by John Lee on Wednesday, December 23, 2020 in May 2020.
Tags: Endothelial cells, Jedi-1, neural stem cells, Neurogenesis