Analyzing the Effects of Mutations on the Binding Abilities of Drugs in Multidrug-Resistant Tuberculosis (MDR-TB) Using Computational Simulations
ABSTRACT
Antibiotic-resistant pathogens are a global crisis. Multidrug-Resistant Tuberculosis (MDR-TB) is a strain of TB that infects ~500,000 people per year, inflicting devastation. Ideally, scientists could identify potential resistance mutations before they manifest in patients. Although there are current experimental techniques for identification, they are costly, time-consuming, and specialized. I propose an inexpensive and novel yet easily operatable pipeline for resistance mutation identification. Using protein structure prediction algorithms (AlphaFold) and molecular docking (HADDOCK), I am able to study the effects of different single-nucleotide polymorphisms (SNPs) on the binding affinity of protein-drug combinations. Focusing specifically on the most common protein-drug combination of katG and Isoniazid for MDR-TB, I propose two novel mutations, N323S and N330Q, that exhibit all hallmarks of a resistance mutation. Specifically, both mutated katG structures demonstrate slightly worse binding to Isoniazid, on par with a known benchmark resistance mutation (S315T). A computational model of predicting mutagenesis is important to provide a more cost-effective method of streamlining the discovery of drug-resistant mutations.
INTRODUCTION.
Since the discovery of penicillin in 1928 [1], antibiotics have saved innumerable lives. For 100 years, humanity has enjoyed the luxury of cheap, effective therapeutics for devastating illnesses like Tuberculosis. However, as humans use more antibiotics for common diseases, we create a selective evolutionary pressure for drug resistant bacterial strains, which have resistance mutations that disable the effect of common drugs. According to the WHO, the antimicrobial resistance crisis is one of the top global public health threats, contributing to >4.95 million deaths every year [2].
Tuberculosis (TB) is a disease caused by Mycobacterium tuberculosis. Spread by coughing and sneezing, tuberculosis is easily transmissible. Prior to antibiotics, the mortality rate of tuberculosis was 50%, earning TB the name “white death” [3]. After antibiotics, the mortality rate decreased. Recently, a new strain known as Multidrug-Resistant Tuberculosis (MDR-TB) has become increasingly prevalent. Of the roughly 10 million TB cases per year, around 500,000 of them are MDR-TB, which is increasing at a 3-5% rate per year [4][5]. MDR-TB does not respond to the most common antibiotics. Not everyone has access to rarer antibiotics that MDR-TB is not resistant to. Thus, mortality rate is 20% for MDR-TB compared to 5% for normal TB [6].
The most common treatment for active TB is Isoniazid (INH). Isoniazid is consumed in a pro-drug form. It is subsequently activated by a critical enzyme encoded by the gene katG in TB bacteria, allowing INH to kill the bacteria by inhibiting the production of an essential component in the bacterial cell wall [7]. However, in MDR-TB, various point substitution mutations in the katG enzyme stop Isoniazid activation. One common mutation is S315T, which I decided to focus on as a mutation representative of other resistance mutations. Critically, S315T does not inhibit the other functions of the enzyme besides Isoniazid activation. Thus, the S315T bacteria survives with drug resistance [8].
Scientists are studying drug resistance mutations in diseases like MDR-TB extensively and hope to predict potential resistance mutations before they even evolve in bacteria. However, this is hard:
The current gold standard is in vitro mutagenesis screens. There are two options [9]:
- Recombinant DNA methods are used to randomly and non-specifically mutate genes of choice.
- Expensive precision tools (like CRISPR-Cas9 + Homology Directed Repair) are used to knock-in the desired mutation.
Either way, current methodology is slow, prohibitively expensive, and/or highly specialized.
In contrast, I believe that mutagenesis screening and prediction of drug resistance mutations can be done through high-throughput computational screening. Here, I develop a novel resistance mutation identification system and validate its efficacy by identifying new mutations for katG-Isoniazid in MDR-TB. The system is inexpensive, accurate in modelling interactions, and can be used to optimize more involved in vitro experiments toward the mutations most likely to cause drug resistance.
MATERIALS AND METHODS.
The computational pipeline I propose is as follows. My procedure focuses on the katG-Isoniazid protein-drug combination but can be applied to any similar pairing. First, I conducted active site modeling.
Active Site Modeling.
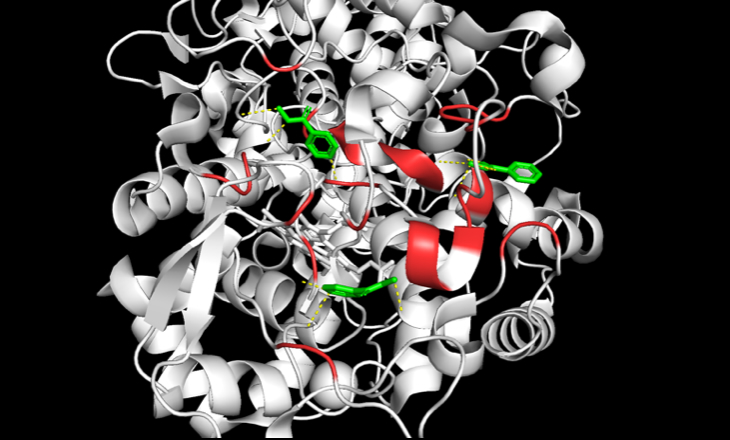
I started with accessing a mirror of the Protein Data Bank (PDB) [10] to retrieve an X-ray Crystallography or Cryo-EM experimental structure for katG. In this case, I used PDB 2CCA. Then, I used PyMol by Schrödinger to identify polar contacts to the literature described active site [11]. Subsequently, I conducted a frequency analysis of known drug resistance mutations to biochemically categorize and estimate the enzymatic effect of each residue (Table S1). Broadly, SNPs can be classified as polar-to-polar, basic-to-polar, etc. based on the nature of the amino acids involved in the mutation. As previously mentioned, current literature also describes the purported active site of katG (Figure 1) [11]. Thus, active site relevant SNPs, particularly ones that modify the biochemical nature of the amino acids, are of particular interest. Using these criteria, I generated a shortlist of potential SNPs not currently described in strains of MDR-TB to test.
AlphaFold Structure Generation.
With the shortlist of mutations to test, the primary amino acid sequence was extracted from the experimentally determined PDB structure in FASTA format. The mutations were then introduced to the FASTA file through editing on a preferred text editor. Finally, the novel mutated structure was determined using AlphaFold. At the time of experimentation, AlphaFold 3 had not been released, and the GitHub release of AlphaFold 2 [12] was used instead.
Molecular Docking Simulation.
Finally, I isolated the Isoniazid ligand from the experimentally determined PDB structure using PyMol. The ligand was then subsequently docked to each mutated structure using High Ambiguity Driven protein-protein Docking (HADDOCK), a flexible docking approach benchmarked with high accuracy for docking interactions relative to other docking methods (Figure 2) [13]. The resultant weighted HADDOCK score can serve as a proxy for binding affinity when considering mutated structures.
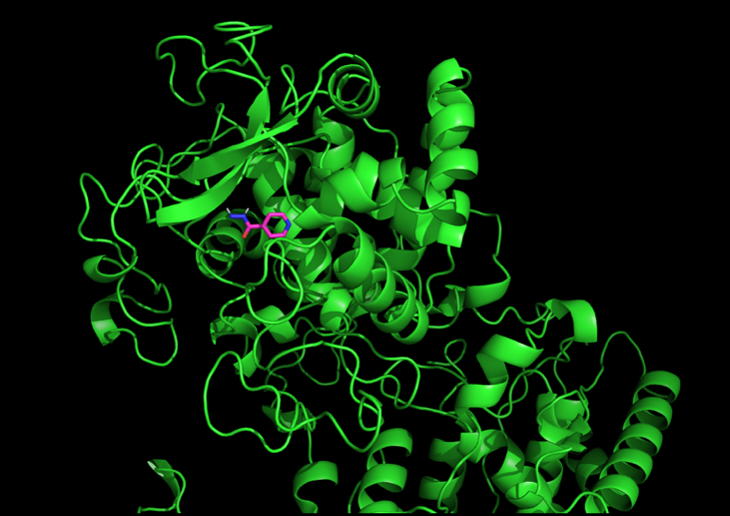
In addition to conducting this procedure for the shortlist of mutations, I also conducted docking for the S315T mutation—a known drug resistance mutation—as a benchmark to compare the mutated HADDOCK scores against.
RESULTS.
Validating Pipeline Accuracy.
Before proceeding to analyze HADDOCK molecular docking scores for variants, I first validated the accuracy of the generated structures by AlphaFold. Root-mean-square-deviation, or RMSD, is a common metric used to determine structural deviation between two protein structures. A low RMSD, measured in angstroms, would imply AlphaFold produced a structure extremely similar to the experimental structure. MatchAlign score is a similar score reported by PyMol. An RMSD <2A is considered good [14].
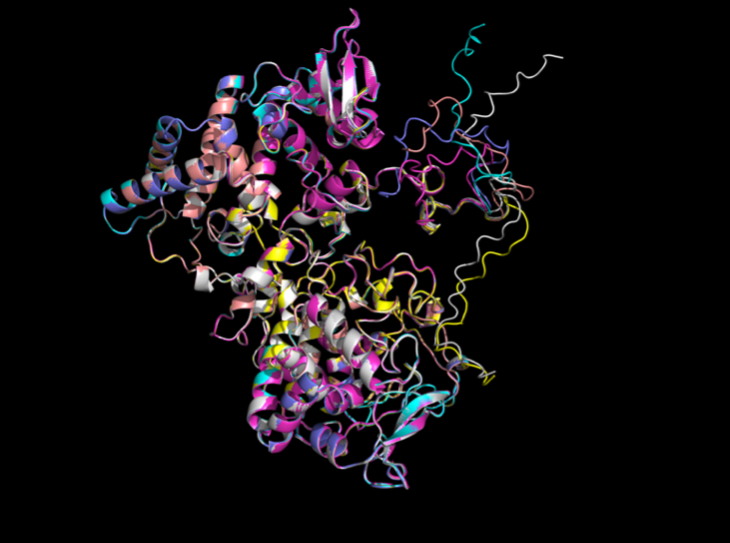
Clearly, the low RMSD values indicate strong structural prediction results from AlphaFold on katG (Table 1). Qualitatively, we can visually see minimal structural deviation, which corresponds to our understanding that SNPs should not broadly change protein structure (Figure 3).
| Table 1. RMSD Between Experimental and AlphaFold Structures | ||
| Structure | RMSD Value (Å) | MatchAlign Score |
| Experimental (PDB) KatG vs. AlphaFold KatG | 0.434 (4593 to 4593 atoms) | 3189.517 |
| Experimental (PDB) KatG w/ S315T Mutation vs. AlphaFold KatG w/ S315T Mutation | 0.526 (4686 to 4686 atoms) | 3132.780 |
Finally, knowing that predicted HADDOCK scores docked against Isoniazid should not deviate too much between different SNP mutations, I compared the AlphaFold structures to PyMol’s simple amino acid substitution tool (Table 2). HADDOCK scores are represented in HADDOCK score units, which is a linear combination of various energies and buried surface area [13]. There is no absolute score which represents ‘good’ docking. The score must be compared relatively. On average, AlphaFold deviated only 5.58 HADDOCK score units, while PyMol’s tool deviated 9.52 from the experimentally derived control S315T mutation. Thus, AlphaFold is more accurate for docking.
| Table 2. HADDOCK Score Difference to S315T PDB’s Docking Score | ||
| Variant/Mutation | AlphaFold Score Difference | PyMol Score Difference |
| Variant 1: T314S | 1.7 | 12.5 |
| Variant 2: N323Q | 2.9 | 6.2 |
| Variant 3: N323S | 9.5 | 9.1 |
| Variant 4: N330Q | 12.4 | 9.0 |
| Variant 5: N330S | 1.4 | 10.8 |
| Average | 5.58 | 9.52 |
Determining Novel Resistance Mutation for katG.
Table 3 reports HADDOCK values of the shortlisted mutations/variants tested. Additional metrics beyond the HADDOCK score are reported in Table S2.
| Table 3. HADDOCK Values for All Mutations | |||
| Variant/Mutation | Structure Origin | Mutation Biochemistry | HADDOCK Score |
| Wild Type | RCSB Protein Data Bank | N/A | 292.8 +/- 7.9 |
| Wild Type | AlphaFold | N/A | 289.5 +/- 5.2 |
| Mutation: S315T | RCSB Protein Data Bank | Polar to Polar | 285.6 +/- 28.8 |
| Mutation: S315T | AlphaFold | Polar to Polar | 291 +/- 4.3 |
| Variant 1: T314S | AlphaFold | Polar to Polar | 287.3 +/- 35.3 |
| Variant 2: N323Q | AlphaFold | Polar to Polar | 282.7 +/- 26.6 |
| Variant 3: N323S | AlphaFold | Polar to Polar | 295.1 +/- 5.0 |
| Variant 4: N330Q | AlphaFold | Polar to Polar | 298.0 +/- 5.0 |
| Variant 5: N330S | AlphaFold | Polar to Polar | 284.2 +/- 28.9 |
DISCUSSION.
As demonstrated by the reported HADDOCK scores for the S315T mutation, which is a known drug resistance mutation, there are a number of criteria we are looking for when analyzing whether a candidate mutation might confer drug resistance.
First, resistance mutations create slightly lower binding interactions between Isoniazid and katG. Higher HADDOCK score approximates lower binding interaction. Specifically, S315T (291.0 score) > Wild Type (289.5 score).
Second, resistance mutations do not create significant binding deficiencies. Otherwise, katG cannot function normally and the MDR-TB bacteria dies. Thus, we cannot expect the scores to be drastically different, or else the entire nature of the protein-drug interaction may be compromised.
There were two katG mutations we screened that satisfied the above criteria. Thus, they are potential resistance mutations within katG. These mutations have never been described in literature. The first mutation is variant 3: N323S. The HADDOCK score was 295.1 Clearly, 295.1 is greater than 289.5. However, it is not significantly greater. This indicates worse binding for the drug Isoniazid and satisfies the two criteria described.
The second mutation is variant 4: N330Q. The HADDOCK score was 298.0. Again, 298.0 is greater than 289.5. However, it is not significantly greater. This indicates worse binding for the drug Isoniazid and satisfies the two criteria described.
The implications of discovering these two mutations are significant. However, computational prediction alone is not sufficient to definitively quantify the variants’ binding affinity—it is instead a preliminary screening measure that indicates these two variants are worth further investigation. Thus, the next step would be to experimentally confirm the binding affinities of the two variants I identified using an in vitro binding assay. Similarly, I hope to also verify in vitro whether these variants exhibit resistance to Isoniazid.
This simple computational pipeline easily replicable for scientists empowers high-throughput analysis of potentially dangerous mutations. With this technology, researchers can model hundreds of mutations in MDR-TB and other diseases might be dangerous in hours and implement necessary interventions or surveillance to address them accordingly after experimental verification. This would be a powerful first step toward solving the drug resistance crisis that threatens our future.
While the results of this research and the discovery of two resistance mutations is certainly exciting, there are several future research directions I would like to take. Specifically, I hope to increase the number of synthetic variants screened by expanding outside of relatively simple active site hotspots by examining the likelihood of mutagenesis at certain residues. In addition, I hope to apply my procedure to other drug resistance pathogens, like Vancomycin-resistant Enterococcus or C. difficile. Greater accuracy can be ascertained by incorporating other docking pipelines and protein generation algorithms, such as AutoDock Vina and RosettaFold respectively. Lastly, I hope to incorporate some form of machine learning to better identify mutations to test. A relatively simple supervised MLP with mutations, protein structure data, and whether they ultimately were resistance mutations can be envisioned to predict whether new mutations are likely to cause resistance.
CONCLUSION.
Using a simple yet powerful computational pipeline, I was able to identify two new potential resistance mutations for Multidrug-Resistant Tuberculosis (MDR-TB). The pipeline is high-throughput and easily applicable to other diseases of interest. In comparison to in vitro mutagenesis screens, this procedure costs less, requires less time, and is easy to operate. This represents not only an increase in understanding of how MDR-TB evolve in the future, but also a new tool to further elucidate drug resistance mechanisms for all diseases.
ACKNOWLEDGMENTS.
I would like to thank Mr. Jason Lee from Lynbrook High School for his support in the project.
SUPPORTING INFORMATION.
Supporting information includes frequency analysis data from the active site modeling procedure and full HADDOCK run data for all katG mutations.
REFERENCES.
- A. Fleming, On the Antibacterial Action of Cultures of a Penicillium, with Special Reference to their Use in the Isolation of B. influenzæ. Br J Exp Pathol 10, 226–236 (1929).
- 2. J. Murray et al., Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. The Lancet. 399, 629–655 (2022).
- G. Mancuso, A. Midiri, S. De Gaetano, E. Ponzo, C. Biondo, Tackling drug-resistant tuberculosis: New challenges from the old pathogen mycobacterium tuberculosis. Microorganisms. 11, 2277 (2023).
- S. Tiberi et al., Accelerating development of new shorter TB treatment regimens in anticipation of a resurgence of multi-drug resistant TB due to the COVID-19 pandemic. International Journal of Infectious Diseases 113, S96-S99 (2021).
- E. A. Kendall, M. O. Fofana, D. W. Dowdy, Burden of transmitted multidrug resistance in epidemics of tuberculosis: A transmission modelling analysis. The Lancet Respiratory Medicine 3, 963–972 (2015).
- E. Kizito et al., Risk factors for mortality among patients diagnosed with multi-drug resistant tuberculosis in uganda: A case-control study. BMC Infectious Diseases 21, 292 (2021).
- C. O’Connor, P. Patel, M. F. Brady, Isoniazid. StatPearls [Internet] (Treasure Island (FL): StatPearls Publishing, 2024).
- S. Yu, S. Girotto, C. Lee, R. S. Magliozzo, Reduced affinity for isoniazid in the S315T mutant of mycobacterium tuberculosis katg is a key factor in antibiotic resistance. Journal of Biological Chemistry 278, 14769–14775 (2003).
- A. Zimmermann, J. E. Prieto-Vivas, K. Voordeckers, C. Bi, K. J. Verstrepen, Mutagenesis techniques for evolutionary engineering of microbes – exploiting CRISPR-cas, oligonucleotides, recombinases, and polymerases. Trends in Microbiology 32, 884–901 (2024).
- S. K. Burley et al., RCSB Protein Data Bank: Biological Macromolecular Structures Enabling Research and education in Fundamental Biology, biomedicine, biotechnology and Energy. Nucleic Acids Research 47, D464-D474 (2018).
- S. M. Kapetanaki, X. Zhao, S. Yu, R. S. Magliozzo, J. P. M. Schelvis, Modification of the active site of mycobacterium tuberculosis katg after disruption of the met–tyr–trp cross-linked adduct. Journal of Inorganic Biochemistry 101, 422–433 (2007).
- J. Jumper et al., Highly accurate protein structure prediction with alphafold. Nature 596, 583–589 (2021).
- C. Dominguez, R. Boelens, A. M. Bonvin, Haddock: A protein−protein docking approach based on biochemical or biophysical information. Journal of the American Chemical Society 125, 1731–1737 (2003).
- O. Carugo, S. Pongor, A normalized root‐mean‐square distance for comparing protein three‐dimensional structures. Protein Science 10, 1470–1473 (2001).
Posted by buchanle on Friday, June 20, 2025 in May 2025.
Tags: Drug Resistance, MDR-TB, Molecular Docking, Protein Folding

