Nuclear Virus-Host Interactions During Coronavirus Infection
ABSTRACT
Coronaviruses are a group of enveloped RNA viruses that are of increasing public health concern, especially in light of recent outbreaks. Like almost all RNA viruses, coronaviruses replicate primarily in the cytoplasm; therefore, the incentive for coronaviruses to target the nucleus is not obvious. Recent research, however, has revealed that coronaviruses commonly employ sophisticated strategies to subvert cellular processes in and around the nucleus in order to promote viral replication. These nuclear virus-host interactions have allowed for coronaviruses to interfere with nuclear transport, perturb nucleolar functions, and regulate host gene expression during infection. An improved understanding of these interactions not only opens avenues for the development of novel therapeutic strategies, but also serves to improve our broader understanding of cell and coronavirus biology.
INTRODUCTION.
Coronaviruses are a group of enveloped viruses with single-stranded, positive-sense RNA genomes. In addition to causing diseases in a variety of animal species, including several economically significant vertebrates (e.g. pigs and chickens), there are seven coronaviruses that can infect humans, five of which have only been identified since 2002 [1]. Consistent with coronaviruses’ general prevalence among domestic mammals and rodents, all human coronaviruses (HCoVs) are believed to have zoonotic origins. Evidence also suggests that all HCoVs originated in bats before spilling over to human hosts, likely circulating in intermediate animal hosts before adapting to infect humans [2,3].
Of the seven HCoVs, four (HCoV-OC43, HCoV-HKU1, HCoV-229E, and HCoV-NL63) produce mild, cold-like symptoms and are generally self-limiting, though in rare cases, these viruses can cause severe lower respiratory complications in elderly, infant, or immunocompromised patients [4, 5]. Collectively, these four endemic coronaviruses are responsible for 15-30% of all cases of the common cold in humans [6]. In contrast, the three other HCoVs (SARS-CoV-1, MERS-CoV, and SARS-CoV-2) are highly pathogenic, are associated with significant mortality rates, and are responsible for costly recent outbreaks [1]. Even prior to the COVID-19 pandemic, the high transmissibility of many HCoVs and the severe diseases associated with the SARS-CoV-1 and MERS-CoV viruses were cause for major public health concern. The emergence of SARS-CoV-2, which possesses both a very high transmissibility and a significant case fatality rate, cemented the need for a thorough understanding of the pathogenesis of coronaviruses and of the cellular mechanisms behind successful coronavirus replication.
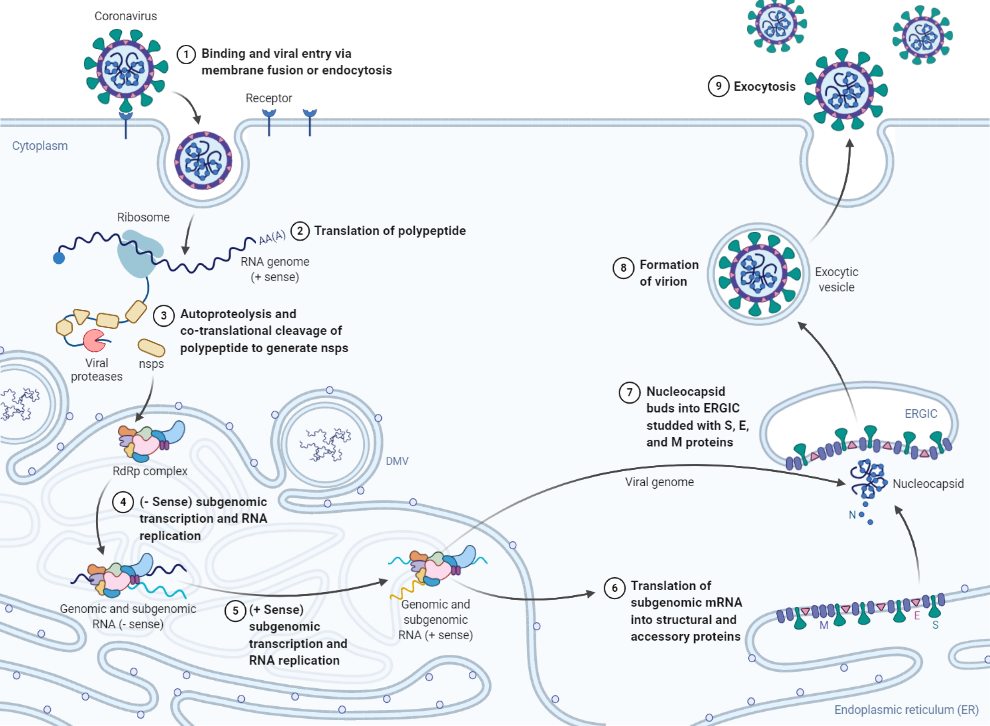
Broadly speaking, viruses are obligate parasites, relying on the resources of the host cell to proliferate. This obligate nature is exemplified by RNA viruses like coronaviruses, which tend to have small genomes and are thus more reliant on interactions with host cell proteins and cellular structures to replicate [6]. Following attachment and entry, the coronavirus replication cycle occurs primarily in the cytoplasm, endoplasmic reticulum (ER), and the ER-Golgi intermediate compartment. The translation and cleavage of viral proteins, the replication of the viral genome, and the assembly of structural viral proteins into new virus particles all occur in these regions (Figure 1) [1]. Accordingly, much focus has been devoted to investigating the intricate processes involved in viral replication that occur in these cellular regions, as the role of virus-host interactions in other subcellular regions is not obvious.
Figure 1. Coronavirus replication cycle. (Definitions: nsp = nonstructural protein, RdRp = RNA-dependent RNA polymerase, DMV = double-membrane vesicle, ERGIC = endoplasmic-reticulum–Golgi intermediate compartment, S = viral spike protein, M = viral membrane protein, E = viral envelope protein, N = viral nucleocapsid protein). *(Fig 1 BioRender.com template is adapted from J. Biol. Chem.(2020) 295(37) 12910–12934.).
As discussed in the present Review, however, coronaviruses have also found ways to subvert other cellular processes surrounding the nucleus to promote viral proliferation. Here, I will discuss such mechanisms, aiming to illustrate the significant, but less obvious role nuclear interactions possess in coronavirus infection.
NUCLEAR TRANSPORT INTERFERENCE.
Even in the absence of an infection, both nuclear import and export play an integral role in eukaryotic cells. The transport of transcription factors and the export of RNA across the nuclear membrane have cascading effects on the physiology of cells, as well as their responses to certain stimuli [7]. Given the foundational role of nuclear transport, the incentive for viruses to target this process becomes apparent.
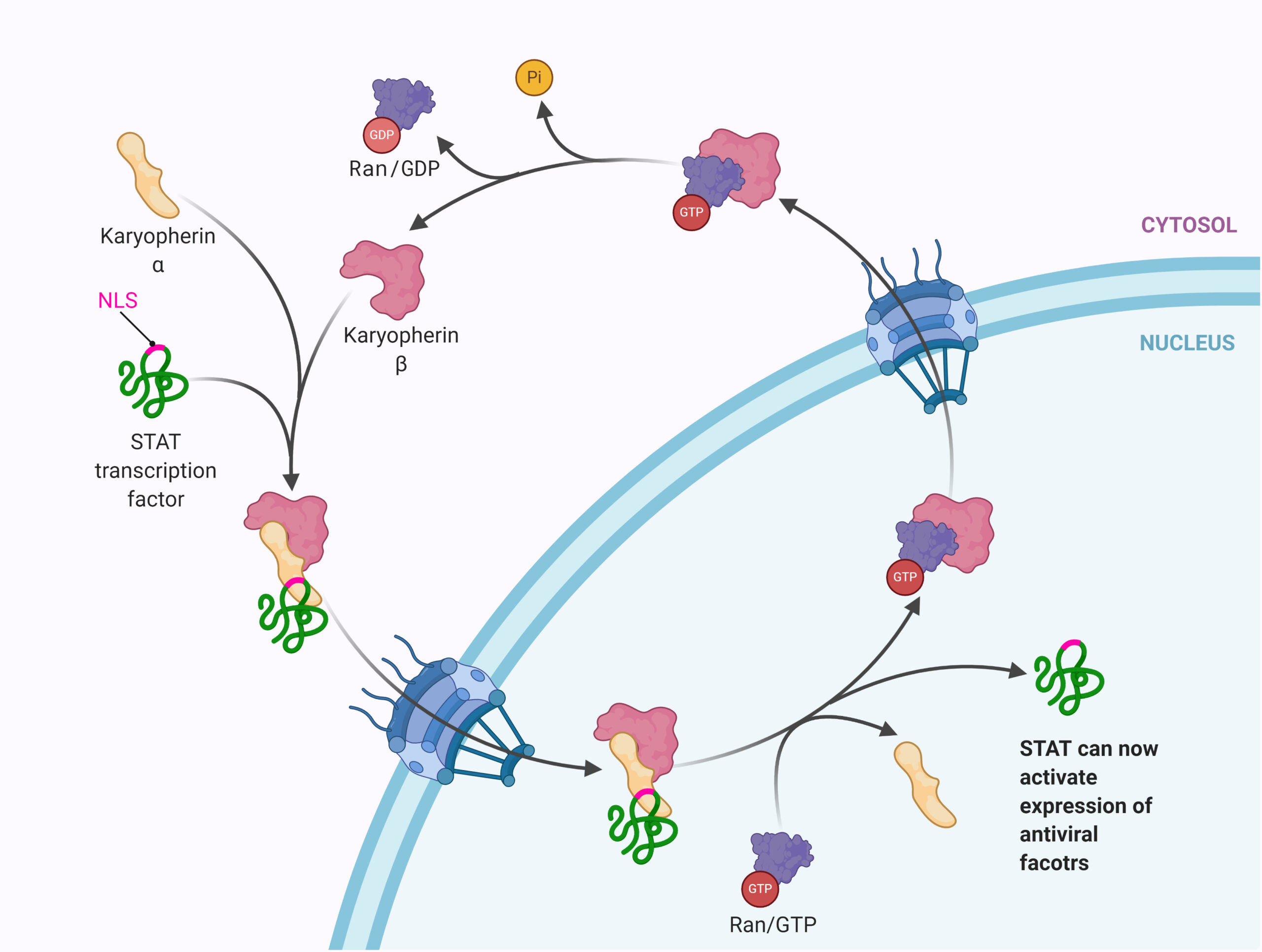
Nuclear pore complexes (NPCs) are the only channels through which macromolecules are able to move from the cytoplasm to the nucleus, or vice-versa. In order to be transported through these pores, macromolecules (proteins, RNA) must be associated with a nuclear transport receptor. Among these nuclear transport receptors are a group of proteins known as karyopherins, which can act as both importins (aiding in nuclear import) or exportins (aiding in nuclear export). As the existing literature reveals, karyopherins are common targets among coronaviruses during infection.
In a 2007 study, Kopecky-Bromberg et al. demonstrated that SARS-CoV-1 accessory protein 6 (ORF6) blocked the translocation of STAT1, a transcription factor significant to the cellular immune response, to the nucleus. The study also found that ORF6 did not prevent STAT1 phosphorylation or dimerization from occurring, indicating that some other novel mechanism outside of a direct modulation of the protein was at play [8]. In a follow-up study by the same research group, this novel mechanism was explored.
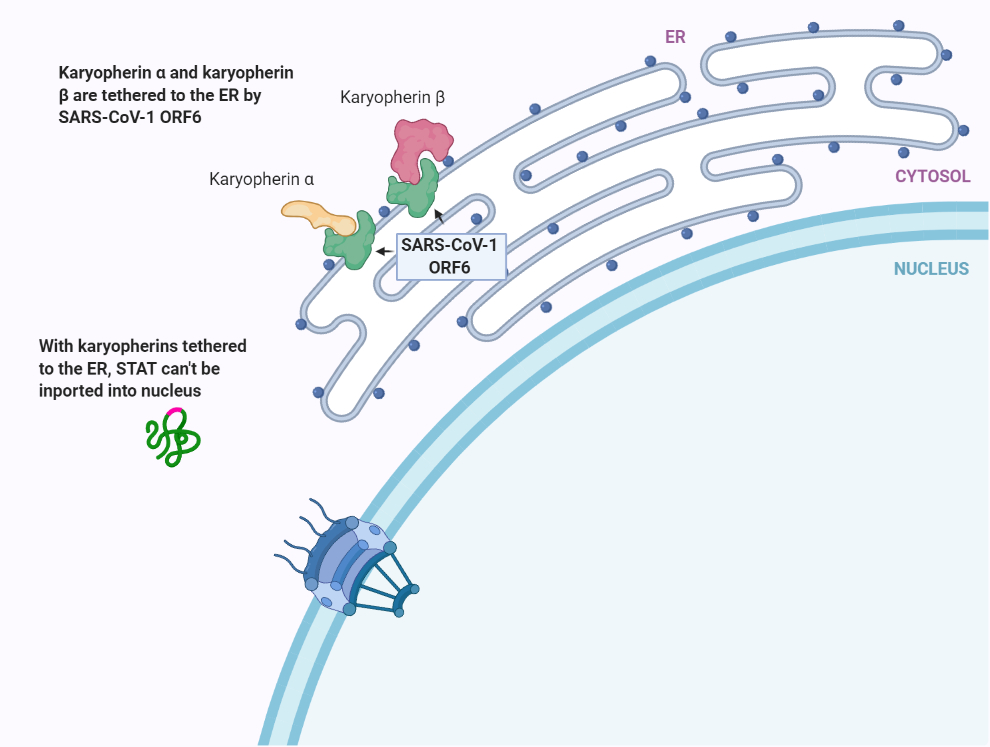
Using a multi-method approach, the group was able to demonstrate that SARS-CoV-1 ORF6 binds to karyopherin alpha 2 and karyopherin beta 1 while also localizing to the ER/Golgi membrane; this allows for ORF6 to tether the two karyopherins to the ER/Golgi membranes, where they are unable to form nuclear import complexes necessary for transport across NPCs (Figure 2) [9]. In addition to STAT1, a later study found that this mechanism was also successful in attenuating the activity of numerous other transcription factors vital to the cellular antiviral response (VDR, CREB1, SMAD4, p53, EpasI, and Oct3/4) [10].
Figure 2. (a) Normal karyopherin-mediated nuclear transport. (b) SARS-CoV-1 ORF6 binds to karyopherin alpha 2 and karyopherin beta 1 and tethers the proteins to the endoplasmic reticulum. This inhibits nuclear transport of transcription factors. *(Fig 2 BioRender.com template is adapted from submission by Mr. Joshua Patrick).
Similarly, SARS-CoV-2 ORF6 also blocks karyopherin-mediated nuclear import of the STAT1 and STAT2 transcription factors. With SARS-CoV-2 however, ORF6 employs a mechanism less direct to karyopherins to accomplish this nuclear import blockage [11].
While the inhibitory effect SARS-CoV-2 ORF6 has on karyopherin-mediated nuclear import is similar to that of SARS-CoV-1 ORF6, the subcellular localization of SARS-CoV-2 ORF6 differs notably. As demonstrated by Miorin et al., instead of the ER/Golgi membrane, SARS-CoV-2 ORF6 localizes to cells’ NPCs. At the NPCs, SARS-CoV-2 ORF6 was found to interact with the Nup98-Rae1 complex. This complex consists of a nucleoporin component (Nup98) that partially makes up NPCs and an mRNA export factor (Rae1). By interacting with this complex, ORF6 is able to prevent the docking of karyopherins at NPCs, consequently impairing the transport of karyopherin cargo (e.g. STAT1 and STAT2 transcription factors) across the nuclear membrane. This study also found that like SARS-CoV-1, SARS-CoV-2 ORF6 also strongly interacts with karyopherin alpha 2, as well as with karyopherin alpha 1. However, modifying SARS-CoV-2 ORF6 to prevent its interaction with the Nup98-Rae1 but conserve its interactions with karyopherins almost completely removed the viral protein’s inhibitory effect on nuclear transport, indicating that the interactions with karyopherins are not responsible for the inhibitory effect SARS-CoV-2 ORF6 has on nuclear transport [11].
Multiple other coronavirus proteins have been found to also interact with host proteins involved in nuclear transport processes [12-14], though the functions and mechanisms of these interactions have not been thoroughly elucidated.
NUCLEOLAR INTERACTIONS.
Within the nuclear envelope, the nucleolus takes on a dynamic and vital role. In addition to being the site of ribosome biogenesis, the nucleolus is now understood to participate in a multitude of other essential cellular processes, including cell cycle regulation, stress response mediation, and signal recognition particle assembly. The protein-protein and protein-nucleic acid interactions that make up the nucleolus are, thus, constantly changing in response to the condition of the cell [6].
Corresponding with the nucleolus’ increasingly dynamic profile, an emerging paradigm over recent decades is the targeting of this subnuclear structure by RNA viruses [6]. Despite replicating in the cytoplasm, numerous RNA viruses, including some coronaviruses, have evolved to target the multifunctional nucleolus, consequently helping facilitate viral replication. Here, I will discuss such interactions involving coronavirus proteins.
The nucleolar phosphoprotein B23 is involved in a plethora of significant nucleolar functions. In addition to binding to single- and double-stranded nucleic acids, B23 also acts as a histone chaperone, participates in ribosomal biogenesis and export, promotes genomic stability, acts as an endoribonuclease, and plays a critical role in centrosome duplication. The expression of B23 also is generally proportional with cell growth rate, indicating that it possesses a positive role in cell growth and proliferation [15]. Given the versatility of B23, it is not surprising that the phosphoprotein interacts with a number of different viruses, having widely varying effects on the respective viruses’ abilities to replicate [16-20]. Among these B23-interacting viruses is the coronavirus SARS-CoV-1 [21].
SARS-CoV-1 nucleocapsid protein (N) is a multifunctional protein that plays a critical role in SARS-CoV-1 infection, regulating many cellular pathways and facilitating both viral packaging and viral core formation [22]. SARS-CoV-1 N has also consistently been found to induce cell cycle arrest in infected cells to help promote replication [22, 23]. It is believed that SARS-CoV-1 N is able to accomplish this through an interaction with B23 [21].
Both in vivo and in vitro, a strong association forms between SARS-CoV-1 N and B23. Interestingly, the location of the B23-binding domain on SARS-CoV-1 N indicates that the interaction does not function to import SARS-CoV-1 N into the nucleus, which is the typical function of other viral protein-B23 interactions. Rather, SARS-CoV-1 N binds to B23 to inhibit the phosphorylation of the nucleolar protein [21]. Since the phosphorylation of B23 is necessary for the protein to dissociate with centrosomes, and since the dissociation of centrosomal B23 is necessary for the cell cycle to progress [24], it is hypothesized that the cell cycle arrest induced by SARS-CoV-1 N is achieved through this interaction with B23 [21].
In addition to the virus’ nucleocapsid protein, SARS-CoV-1 accessory protein 3b (ORF3b) also predominantly localizes to the nucleolus [25], though the functional purpose of this localization is ambiguous. For example, overexpression of SARS-CoV-1 ORF3b has been found to notably induce both apoptosis and necrosis in vitro, though the localization of ORF3b to the nucleolus is not important to activating these cell-death pathways [26]. Further, SARS-CoV-1 ORF3b also modulates the activity of host transcriptional factor RUNX1b, with a hypothesized relevance in upregulating cytokine and chemokine levels, though this interaction was indicated to occur in the extra nucleolar space of the nucleus [27]. Therefore, additional work is necessary in order to fully explain the function of SARS-CoV-1 ORF3b’s localization to the nucleolus.
Along with SARS-CoV-1 N, multiple nucleolar interactions have been confirmed involving the avian coronavirus/infectious bronchitis virus (IBV) nucleocapsid protein. IBV N, which is functionally similar to SARS-CoV-1 N [28], has been found to interact with both nucleolin, a nucleolar protein involved extensively in transcription, ribosome assembly, and mRNA translation and stability [29], and fibrillarin, a protein involved in the processing of ribosomal RNA and cell cycle regulation [30, 31]. It is through these interactions that IBV N is able to alter the localization of fibrillarin. Although the precise mechanism behind this effect is unknown, it is hypothesized that this allows IBV N to inhibit host transcription factors and interfere with ribosomal biogenesis, naturally interfering with host gene expression [31].
In IBV infected cells, the tumor suppressor p53 protein, which normally localizes to the nucleolus and nucleus, is redistributed primarily to the cytoplasm. It is believed that IBV N is responsible for this redistribution, as IBV N was found to co-localize with p53 both in the nucleus/nucleolus (although not significantly) and in the cytoplasm with other IBV proteins [32]. IBV N has been shown to significantly reduce cell growth [32, 33], and the redistribution of p53 is likely what accounts for this [32].
Porcine epidemic diarrhea coronavirus (PEDV) is another coronavirus whose nucleocapsid protein localizes to nucleolus. PEDV N has a confirmed interaction with nucleolar phosphoprotein nucleophosmin (NPM1), and this interaction was found to protect NPM1 from proteolytic degradation, which in turn promotes cell survival [34]. Programmed cell death is a common strategy employed by hosts to prevent successful viral replication [35], and the PEDV N-NPM1 interaction works to evade this strategy.
The nucleocapsid proteins of murine hepatitis coronavirus (MHV) and transmissible gastroenteritis coronavirus (TGEV) also localize to the nucleolus. Although nucleolar protein interactions involving either MHV N or TGEV N have yet to be confirmed, TGEV N and MHV N are both indicated to induce cell cycle arrest/delay [33], similar to SARS-CoV-1 N and IBV N. Regardless, the localizations of MHV N and TGEV N help further cement that the nucleolus is commonly targeted during coronavirus infection, especially by viral nucleocapsid proteins.
HOST GENE REGULATION.
As hosts’ first line of defense, the innate immune response plays a pivotal role in determining a host’s ability to curb viral infections. At the cellular level, a key component of the innate immune response is the effective interferon-mediated response [36]. Interferons are signaling proteins released by host cells in response to a viral infection; by interacting with the receptors of nearby cells, interferons are able to activate transcription factors that heighten the expression of genes encoding antiviral defenses. These genes are known as interferon-stimulated genes (ISGs). In order to facilitate replication, coronaviruses employ numerous strategies to suppress the expression of these host genes.
Indeed, some of the mechanisms already discussed in this Review could be viewed as host gene regulation mechanisms. As discussed above, by interfering with nuclear transport during infection, some coronaviruses are able to prevent STAT transcription factors from entering the nucleus; in turn, this downregulates the expression of the ISGs that STAT transcription factors activate [9]. By interfering with ribosomal biogenesis in the nucleolus, it is hypothesized that IBV N is also able to have a natural, suppressing effect on host gene expression [31]. In addition to these, coronaviruses have been found to employ other strategies in and around the nucleus to regulate the expression of host genes.
SARS-CoV-2 N, which likely localizes predominantly to the nucleus [37], directly interacts with STAT1 and STAT2 to suppress the phosphorylation of the transcription factors. This disrupts the downstream expression of ISGs [38]. MERS nonstructural protein 1 (nsp1), which also localizes to the nucleus, targets mRNAs transcribed in the nucleus for mRNA degradation and translation inhibition, though spares mRNAs originating in the cytoplasm, such as those of a virus. By doing this, MERS nsp1 is able to promote the expression of viral genes while suppressing the expression of host genes [39].
Evasion of the interferon-mediated response, with the larger goal of suppressing host gene expression, is achieved by coronaviruses through a number of strategies in other, non-nuclear regions of the cell [40]. These interactions and mechanisms, however, are not within the scope of the present Review.
CONCLUSION.
Largely due to recent outbreaks, coronaviruses have taken on a heightened level of public health concern. Accordingly, significant efforts have been made in recent years to understand the cellular mechanisms behind successful coronavirus replication.
Despite replicating in the cytoplasm, coronaviruses have been revealed to employ numerous mechanisms in and around the nucleus to subvert cellular processes and facilitate viral replication. Increasingly so, these nuclear virus-host interactions are understood to play significant roles in the larger replication cycle of coronaviruses. Increased knowledge of this aspect of coronavirus infection holds the potential to not only facilitate the development of novel therapeutic strategies, but also further our broader understanding of cell and coronavirus biology.
ACKNOWLEDGMENTS.
I would like to thank the Plate Research Group, Dr. Lars Plate, and the School for Science and Math at Vanderbilt. I would also like to thank Jonathan Davies for his continued guidance.
REFERENCES.
- Z. W. Ye, S. Yuan, K. S. Yuen, S. Y. Fung, C. P. Chan, D. Y. Jin, Zoonotic origins of human coronaviruses. International Journal of Biological Sciences 16, 1686-1697 (2020).
- S. Su, G. Wong, W. Shi, J. Liu, A. C.K. Lai, J. Zhou, W. Liu, Y. Bi, G. F. Gao, Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends in Microbiology 24, 490-502 (2016).
- G. J. Gorse, T. Z. O’Connor, S.L. Hall, J. N. Vitale, K. L. Nichol, Human coronavirus and acute respiratory illness in older adults with chronic obstructive pulmonary disease. Journal of Infectious Diseases 199, 847-857 (2009).
- F. Pene, A. Merlat, A. Vabret, F. Rozenberg, A. Buzyn, F. Dreyfus, A. Cariou, F. Freymuth, P. Lebun, Coronavirus 229E-related pneumonia in immunocompromised patients. Clinical Infectious Diseases 37, 929-932 (2003).
- Y. Lim, Y. Ng, J. Tam, D. Liu, Human coronaviruses: a review of virus-host interactions. Diseases 4, 26 (2016).
- J. Hiscox, RNA viruses: Hijacking the dynamic nucleolus. Nature Reviews Microbiology 5, 119–127 (2007).
- G. M. Cooper, “The Nuclear Envelope and Traffic between the Nucleus and Cytoplasm” in The Cell: A Molecular Approach. (Sinauer Associates, Sunderland, MA, ed. 2, 2000).
- S. Kopecky-Bromberg, L. Martinez-Sobrido, M. Frieman, R. A. Baric, P. Palese, Severe Acute Respiratory Syndrome Coronavirus Open Reading Frame (ORF) 3b, ORF 6, and Nucleocapsid Proteins Function as Interferon Antagonists. Journal of Virology 81, 548-557 (2007).
- M. Frieman, B. Yount, M. Heise, S. Kopecky-Bromberg, P. Palese, R. S. Baric, Severe Acute Respiratory Syndrome Coronavirus ORF6 Antagonizes STAT1 Function by Sequestering Nuclear Import Factors on the Rough Endoplasmic Reticulum/Golgi Membrane. Journal of Virology 81, 9812-9824 (2007)
- A. C. Sims, S. C. Tilton, V. D. Menachery, L. E. Gralinski, A. Schafer, M. M. Matzke, B.-J. M. Webb-Robertson, J. Chang, M. L. Luna, C. E. Long, A. K. Shukla, A. R. Bankhead, S. E. Burkett, G. Zornetzer, C.-T. K. Tseng, T. O. Metz, R. Pickles, S. McWeeney, R. D. Smith, M. G. Katze, K. M. Waters, R. S. Baric, Release of Severe Acute Respiratory Syndrome Coronavirus Nuclear Import Block Enhances Host Transcription in Human Lung Cells. Journal of Virology 87, 3885-3902 (2013).
- L. Miorin, T. Kehrer, M. T. Sanchez-Aparicio, K. Zhang, P. Cohen, R. S. Patel, A. Cupic, T. Makio, M. Mei, E. Moreno, O. Danziger, K. M. White, R. Rathnasinghe, M. Uccellini, S. Gao, T. Aydillo, I. Mena, X. Yin, L. Martin-Sancho, N. J. Krogan, S. K. Chanda, M. Schotsaert, R. W. Wozniak, Y. Ren, B. R. Rosenberg, B. M. A. Fontoura, A. García-Sastre, SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proceedings of the National Academy of Sciences 117, 28344-28354 (2020).
- F. Messina, E. Giombini, C. Agrati, F. Vairo, T. A. Bartoli, S. Al Moghazi, M. Piacentini, F. Locatelli, G. Kobinger, M. Maeurer, A. Zumla, M. R. Capobianchi, F. N. Lauria, G. Ippolito, COVID-19: viral–host interactome analyzed by network based-approach model to study pathogenesis of SARS-CoV-2 infection. Journal of Translational Medicine 18, 233, (2020).
- Y. Terada, K. Kawachi, Y. Matsuura, W. Kamitani, MERS coronavirus nsp1 participates in an efficient propagation through a specific interaction with viral RNA. Virology 511, 95-105 (2017).
- M. H. Uddin, J. A. Zonder, A. S. Azmi, Exportin 1 inhibition as antiviral therapy. Drug Discovery Today 25, 1775-1781 (2020).
- M. Okuwaki, The structure and functions of NPM1/Nucleophsmin/B23, a multifunctional nucleolar acidic protein. Journal of Biochemistry 143, 441-448 (2008).
- Y. Tsuda, Y. Mori, T. Abe, T. Yamashita, T. Okamoto, T. Ichimura, K. Moriishi, Y. Matsuura, Nucleolar protein B23 interacts with Japanese encephalitis virus core protein and participates in viral replication. Microbiology and Immunology 50, 225-234 (2006).
- R.T. Mai, T.S. Yeh, C.F. Kao, S.K. Sun, H.H. Huang, Y.H. W. Lee, Hepatitis C virus core protein recruits nucleolar phosphoprotein B23 and coactivator p300 to relieve the repression effect of transcriptional factor YY1 on B23 gene expression. Oncogene 25, 448-462 (2006).
- A.M. Passos-Casthilo, C. Marchand, D. Archambault, B23/nucleophosmin interacts with bovine immunodeficiency virus Rev protein and facilitates viral replication. Virology 515, 158-164 (2018).
- R. Abraham, S. Singh, S. R. Nair, N. V. Hulyalkar, A. Surendran, A. Jaleel, E. Sreekumar, Nucleophosmin (NPM1)/B23 in the Proteome of Human Astrocytic Cells Restricts Chikungunya Virus Replication. Journal of Proteome Research 16, 4144-4155 (2017).
- W. Mai, F. Huang, H. Chen, Y. Zhou, Y. Chen, Nervous necrosis virus capsid protein exploits nucleolar phosphoprotein Nucleophosmin (B23) function for viral replication. Virus Research 230, 1-6 (2017).
- Y. Zeng, L. Ye, S. Zhu, H. Zheng, P. Zhao, W. Cai, L. Su, Y. She, Z. Wu, The nucleocapsid protein of SARS-associated coronavirus inhibits B23 phosphorylation. Biochemical and Biophysical Research Communications 369, 287-291 (2008).
- M. Surjit, S. K. Lal, “The Nucleocapsid Protein of the SARS Coronavirus: Structure, Function and Therapeutic Potential” in Molecular Biology of the SARS-Coronavirus. (Springer, Berlin, Germany, ed. 1, 2009).
- K.A. Timani, Q. Liao, L. Ye, Y. Zeng, J. Liu, Y. Zheng, L. Ye, X. Yang, K. Lingbao, J. Gao, Y. Zhu, Nuclear/nucleolar localization properties of C-terminal nucleocapsid protein of SARS coronavirus. Virus Research 114, 23-34 (2005).
- M. Okuda, H.F. Horn, P. Tarapore, Y. Tokuyama, A.G. Smulian, P.K. Chan, E.S. Knudsen, I.A. Hofmann, J.D. Snyder, K.E. Bove, K. Fukasawa, Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell 103, 127-140 (2000).
- X. Yuan, Z. Yao, Y. Shan, B. Chen, Z. Yang, J. Wu, Z. Zhao, J. Chen, Y. Cong, Nucleolar localization of non-structural protein 3b, a protein specifically encoded by the severe acute respiratory syndrome coronavirus. Virus Research 114, 70-79 (2005).
- S. Khan, B.C. Fielding, T.H. Tan, C.F. Chou, S. Shen, S.G. Lim, W. Hong, Y.J. Tan, Over-expression of severe acute respiratory syndrome coronavirus 3b protein induces both apoptosis and necrosis in Vero E6 cells. Virus Research 122, 20-27 (2006).
- B. Varshney, S. Agnihotram,Y. Tan, R. Baric, S. K. Lal, SARS Coronavirus 3b Accessory Protein Modulates Transcriptional Activity of RUNX1b. PLOS ONE 7, e29542 (2012).
- R. McBride, M. van Zyl, B. C. Fielding, The Coronavirus Nucleocapsid Is a Multifunctional Protein. Viruses 6, 2991-3018 (2014).
- K. Abdelmohsen, M. Gorospe, RNA-binding protein nucleolin in disease. RNA Biology 9, 799-808 (2012).
- K. Newton, E. Petfalski, D. Tollervey, J. F. Caceres, Fibrillarin Is Essential for Early Development and Required for Accumulation of an Intron-Encoded Small Nucleolar RNA in the Mouse. Molecular and Cellular Biology 23, 8519-8527 (2003).
- H. Chen, T. Wurm, P. Britton, G. Brooks, J.A. Hiscox, Interaction of the coronavirus nucleoprotein with nucleolar antigens and the host cell. Journal of Virology 76, 5233-5250 (2002).
- B.K. Dove, J.H. You, M.L. Reed, S.R. Emmett, G. Brooks, J.A. Hiscox, Changes in nucleolar morphology and proteins during infection with the coronavirus infectious bronchitis virus. Cellular Microbiology 8, 1147-1157 (2006)
- T. Wurm, H. Chen, T. Hodgson, P. Britton, G. Brooks, J.A. Hiscox, Localization to the nucleolus is a common feature of coronavirus nucleoproteins, and the protein may disrupt host cell division. Journal of Virology 75, 9345-9356 (2001).
- D. Shi, H. Shi, D. Sun, J. Chen, X. Zhang, X. Wang, J. Zhang, Z. Ji, J. Liu, L. Cao, X. Zhu, J. Yuan, H. Dong, X. Wang, T. Chang, Y. Liu, L. Feng, Nucleocapsid Interacts with NPM1 and Protects it from Proteolytic Cleavage, Enhancing Cell Survival, and is Involved in PEDV Growth. Scientific Reports 7, 39700 (2017).
- B. J. Thomson, Viruses and apoptosis. International Journal of Experimental Pathology 82, 65-76 (2001).
- Y. Huang, H. Dai, R. Ke, Principles of Effective and Robust Innate Immune Response to Viral Infections: A Multiplex Network Analysis. Frontiers in Immunology 2019, 10:1736 (2019).
- A. B. Gussow, N. Auslander, G. Faure, Y. I. Wolf, F. Zhang, E. V. Koonin, Genomic determinants of pathogenicity in SARS-CoV-2 and other human coronaviruses. PNAS 117, 15193-15199 (2020).
- J. Mu, Y. Fang, Q. Yang, T. Shu, A. Wang, M. Huang, L. Jin, F. Deng, Y. Qiu, X. Zhou, SARS-CoV-2 N protein antagonizes type I interferon signaling by suppressing phosphorylation and nuclear translocation of STAT1 and STAT2. Cell Discovery 6, 65 (2020).
- K. G. Lokugamage, K. Narayanan, K. Nakagawa, K. Terasaki, S. I. Ramirez, C. K. Tseng, S. Makino, Middle East Respiratory Syndrome Coronavirus nsp1 Inhibits Host Gene Expression by Selectively Targeting mRNAs Transcribed in the Nucleus while Sparing mRNAs of Cytoplasmic Origin. Journal of Virology 89, 10970-10981 (2015).
- E. Kindler, V. Thiel, F. Weber, “Interaction of SARS and MERS Coronaviruses with the Antiviral Interferon Response” in vol. 96 of Advances in Virus Research (Academic Press Inc. Cambridge, MA, 2016) 219-243.
Posted by John Lee on Thursday, May 20, 2021 in May 2021.
Tags: Coronavirus, Gene Regulation, Nuclear Transport, Nucleolus, Nucleus