Computational Simulations of Bispecific Aptamers Targeting CD16+ Natural Killer Cells and PD-L1+ Cancer Cells
ABSTRACT
Adoptive cell therapy is an advanced cancer treatment that utilizes natural killer (NK) cells to target and eliminate cancer cells. This therapy leverages the interaction between the PD-L1 receptor on cancer cells and the CD16 receptor on NK cells to enhance immune responses and improve therapeutic outcomes. We hypothesize that the bi-specific aptamer designed in this research will target CD16 and PDL1 receptors, thus inducing NK cell-mediated cancer cell apoptosis. The receptor structure was obtained using AlphaFold 3, an advanced AI tool for predicting 3D protein structures. This research aims to create effective NK cell-based cancer immunotherapies using bispecific aptamers to target and destroy cancer cells. Aptamer modeling was performed using Vfold2D and Vfold3D software to predict secondary and tertiary structures. Molecular docking simulations were conducted using HDOCK 2.0 to study aptamer-receptor interactions. The PLIP web server was used for detailed interaction analysis, identifying bonds and key contacts in the receptor-aptamer complex. The best binder among the aptamers was aptamer C, based on its strong interaction with the predicted binding site and high stability in molecular docking simulations. The study demonstrates the potential of bispecific aptamers targeting CD16 and PD-L1 receptors to enhance NK cell-mediated cancer cell therapy.
INTRODUCTION.
CAR T-cell therapy, known as Chimeric Antigen Receptor T-cell therapy, is a cutting-edge and personalized approach to cancer treatment (Mohanty et al., 2019). It works by modifying a patient’s T cells to target and destroy cancer cells more effectively (Miliotou & Papadopoulou, 2018). The process starts with collecting T cells from the patient’s blood (Vormittag, Gunn, Ghorashian, & Veraitch, 2018). These cells are then altered in a lab to include a particular chimeric antigen receptor (CAR) designed to recognize cancer cells (Sterner & Sterner, 2021). After these modified cells are grown and tested, they are infused into the patient’s bloodstream (Sterner & Sterner, 2021). The CARs help the T cells find and attach to specific markers on the cancer cells, leading to their destruction (Mohanty et al., 2019). This treatment has been highly effective for certain blood cancers, like acute lymphoblastic leukemia and some lymphomas (Anderson & Mehta, 2019). However, it can also cause side effects and challenges, including cytokine release syndrome and neurotoxicity (Anderson & Mehta, 2019).
PD-L1, or programmed death-ligand 1, is a protein found on cell surfaces that helps control how the immune system responds to cells, including cancer cells (Guan, Lim, Mekhail, Chang, & Medicine, 2017). When PD-L1 binds to its receptor PD-1 on T-cells, it stops the T-cells from working, letting cancer cells avoid being attacked by the immune system (Guan et al., 2017). This is a significant way tumors are prevented from being destroyed (Guan et al., 2017). Because of this, PD-L1 is an essential target for cancer treatments (Guan et al., 2017; Hudson, Cross, Jordan-Mahy, & Leyland, 2020). By blocking the PD-1/PD-L1 connection with particular antibodies, T-cells can start working again and attack cancer cells (Guan et al., 2017). This method has shown promise in treating melanoma, lung, and kidney cancers (Hudson et al., 2020).
CD16, also called FcγRIII, is a receptor that plays a vital role in the immune response (Hudson et al., 2020). It’s found on the surface of NK cells, macrophages, and some T cells and helps with antibody-dependent cellular cytotoxicity (ADCC) (Hudson et al., 2020). There are two types of CD16: CD16a and CD16b (Hudson et al., 2020). CD16a is found on NK cells and macrophages, while CD16b is on neutrophils (Ortaldo, Mason, & O’Shea, 1995). These receptors bind to IgG antibodies, causing NK cells to release toxic substances and macrophages to engulf pathogens, helping to get rid of antibody-coated germs and infected or cancerous cells (Ortaldo et al., 1995). Because of its role in ADCC, CD16 is a target for cancer and infectious disease treatments (Ortaldo et al., 1995).
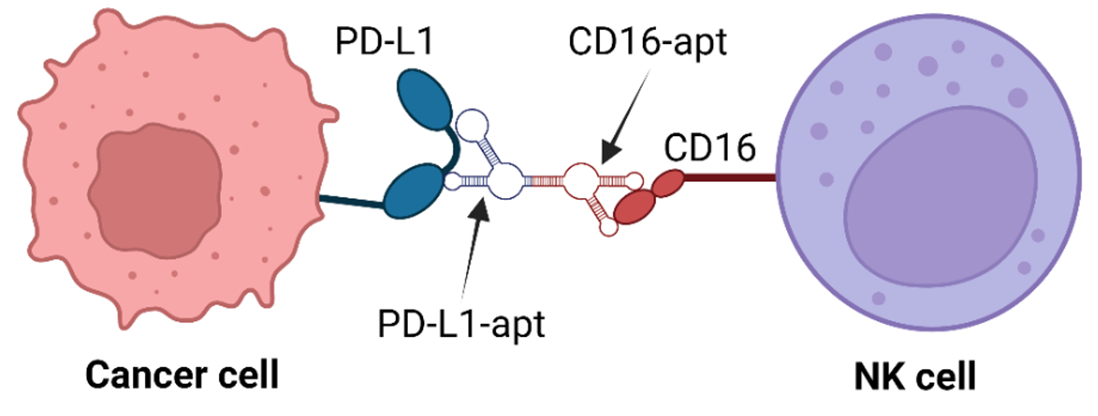
Aptamers are short, single-stranded nucleic acids (DNA or RNA) that can bind to specific target molecules with high affinity and specificity, similar to antibodies (Banerjee & Nilsen-Hamilton, 2013). They are selected through SELEX and can target proteins, small molecules, or cells (Lee, Stovall, & Ellington, 2006). Due to their stability, low immunogenicity, and ease of synthesis, aptamers are increasingly explored for therapeutic and diagnostic purposes (Song, Lee, & Ban, 2012). Bi-specific aptamers are designed to bind two different targets simultaneously, enhancing their precision in targeting cancer cells (Rozenblum, Lopez, Vitullo, & Radrizzani, 2016). Bi-specific aptamers can be engineered for cancer treatment to interact with immune cells, such as NK cells, and cancer-specific proteins, such as PD-L1 (Zheng et al., 2022). This dual-targeting approach enables more effective tumor destruction by boosting the immune system’s response, making them a promising tool for cancer immunotherapy (Zheng et al., 2022).
Recently, Zheng designed bispecific aptamers that bind to both CD16 and PD-L1 receptors on NK and cancer cells, respectively (Zheng et al., 2022). Understanding these bispecific aptamers’ structure and binding mechanism will be crucial for designing novel NK cell-mediated cancer therapy. We hypothesize that the aptamer should bind to a specific binding region of the protein. We have performed computational simulations using molecular docking simulations using HDOCK software to find the CD16-aptamer-PD-L1 complex structure. These results will pave the way for understanding and designing novel therapies against various cancer cells.
MATERIALS AND METHODS.
Receptor Modeling.
In this research, we utilized AlphaFold 3 to predict the 3D structure of proteins, which was refined using molecular modeling tools like PyMOL to remove water molecules, heteroatoms, and non-standard residues (Abramson et al., 2024). AlphaFold 3 is an advanced AI tool that predicts the 3D structure of proteins from their amino acid sequences (Abramson et al., 2024). It’s a massive breakthrough in biology, as knowing the shape of a protein helps scientists understand how it works and interacts with other molecules (Abramson et al., 2024). Amino acid sequences of CD16 and PD-L1 receptors were inputted into AlphaFold 3 to predict their 3D structures. For studying proteins like CD16 and PDL1, AlphaFold 3 gave us a detailed 3D model of their structure, which is essential for studying their role in the immune system.
Aptamer modeling.
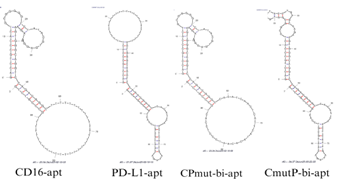
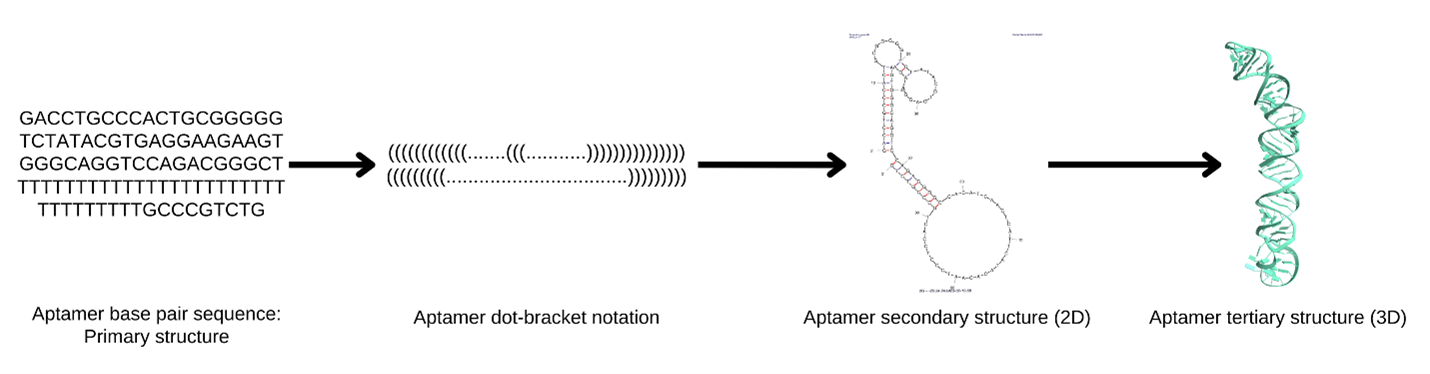
The structural predictions were based on the aptamer’s primary nucleotide sequence (Table 1). Initially, Vfold2D predicted the secondary structure using dot-bracket notation, where dots represent unpaired nucleotides and parentheses denote base pairs, as seen in Figure 1. For example, a simple hairpin structure appears as “…..(((((…..)))))…..” (Xu & Chen, 2021). Once the secondary structure was established, Vfold3D was used to predict and model the tertiary structure (Figure 2), which we then visualized with ChimeraX (Xu & Chen, 2021).
| Table 1. Aptamer sequences used in this research. | |
| A | GACCTGCCCACTGCGGGGGTCTATACGTGAGGAAGAAGTG GGCAGGTCCAGACGGGCTTTTTTTTTTTTTTTTTTTTTTTTTT TTTTTTTGCCCGTCTG((((((((((((…….(((………..)))))))))))))))((((((((( ……………………………))))))))) |
| B | GACCTGCCCACTTTTTTTTTTTTTTTTTTTTTTTTTAGTGGGC AGGTCCAGACGGGCCACATCAACTCATTGATAGACAATGC GTCCACTGCCCGTCTG((((((((((((……………………))))))))))))((((((((( …….((.(((((…..))))).))…..))))))))) |
| C | GACCTGCCCACTGCGGGGGTCTATACGTGAGGAAGAAGTG GGCAGGTCCAGACGGGCCACATCAACTCATTCATACACAA TCCCTCCACTGCCCGTCTG((((((((((((…….(((………..)))))))))))))))( ((((((((……………………………))))))))) |
| D | GACCTGCCCACTCCGGCGGTGTATACCTGAGCAAGAAGTG GGCAGGTCCAGACGGGCCACATCAACTCATTGATAGACAA TGCGTCCACTGCCCGTCTG((((((((((((…(((((….)))…))….))))))))))))(((((((((…….((.(((((…..))))).))…..))))))))) |
Binding site prediction.
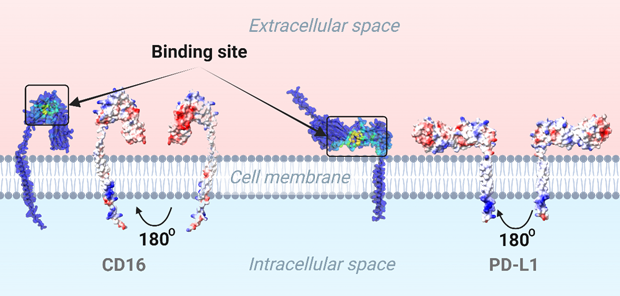
To identify the binding sites, we used GrASP, which predicts potential interaction points on the protein surface. We started using a Graph Neural Network method with the GrASP web server to pinpoint the binding site on the protein’s surface. The 3D structures of CD16 and PD-L1 were inputted to gain the predicted binding regions. As shown in Figure 3, you can see the predicted binding area marked in yellow-green. We hypothesize that the aptamer will specifically attach to this highlighted region. Additionally, we analyzed the electrostatic surface potential (ESP) using ChimeraX, where positively charged areas appear blue, negatively charged regions in red, and neutral areas in white (Figure 3). Since aptamers are highly negative due to phosphate groups, they effectively bind to positively charged sites, aiding in targeted therapy design.
Molecular Docking.
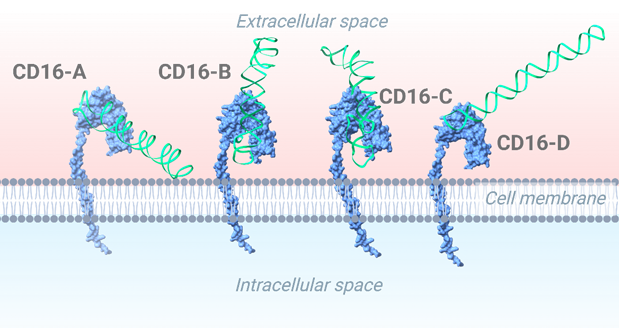
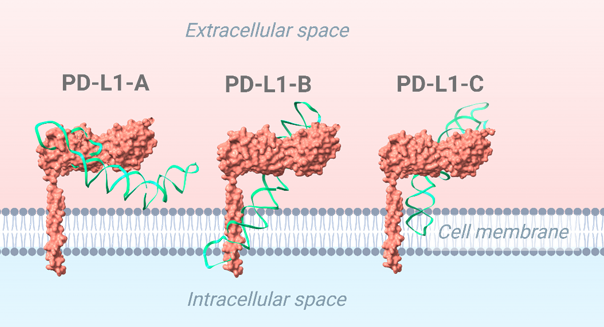
To refine our understanding of aptamer-protein interactions, we performed molecular docking simulations to determine how a small molecule, or ligand, interacts with a target protein or receptor. The receptor and ligand structures were prepared and uploaded to HDock, which docked the aptamer to the target protein (Yan, Zhang, Zhou, Li, & Huang, 2017). The lowest energy structure was selected for further analysis. Specifically, we studied aptamer interactions with CD16/PD-L1 receptors to enhance NK cell-mediated cancer immunotherapy (Figure 4). The docked results are shown in Figures 5 and 6 (Yan, Tao, He, & Huang, 2020). The aptamers were selected based on the following three criteria. We have used the binding site region as an aptamer selection criterion. We specifically selected this region within the aptamer because it strongly connects to the predicted binding site on the receptor, which ChimeraX determined. Using the visual inspection tool on ChimeraX, we confirmed the aptamer’s interaction at the predicted binding site. If the aptamer doesn’t bind through the predicted region, it will result in a weak bond with the receptor since the predicted region is specifically chosen for its stable and more abundant interactions, rendering the aptamer ineffective for its intended purpose. The selected aptamers are shown in Figures 5 and 6.
Data Analysis.
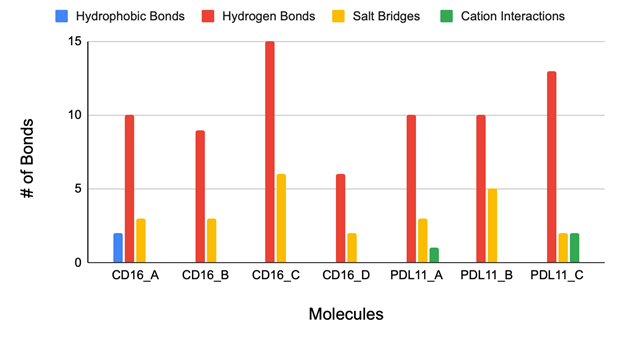
The final receptor-aptamer complexes were analyzed using the PLIP web server, which identified hydrogen bonds, hydrophobic interactions, and other key molecular contacts (Figure 7). The PLIP tool provided detailed information on the specific bonds and interactions between the receptors and aptamers. The results summarized in Figure 7 include hydrogen bonds, hydrophobic interactions, and other key contacts within the receptor-aptamer complex. All the aptamers interact with the binding site of the CD16 protein, as shown in Figure 5. This region is slightly positive, so a negatively-charged aptamer binds to it. Similarly, most the aptamer binds to the druggable site of the PDL1 protein (Figure 6). The central region of the protein is slightly positive (blue).
RESULTS.
Number of Interactions.
The molecular interaction analysis identifies Aptamer C as the most effective bispecific binder for CD16 and PD-L1, demonstrating superior hydrogen bonding and salt bridge formation compared to other candidates. Hydrogen bonds are crucial for molecular recognition and receptor-ligand stability, and Aptamer C forms 15 hydrogen bonds with CD16 and 13 with PD-L1, significantly outpacing the other aptamers. In contrast, Aptamer A forms only 10 hydrogen bonds with both receptors, leading to a weaker binding profile, while Aptamer B forms 9 with CD16 and 10 with PD-L1, indicating lower overall stability. Aptamer D performs the worst, forming just 6 hydrogen bonds with CD16 and none with PD-L1, making it ineffective for dual-receptor targeting. Salt bridges, which provide electrostatic stabilization, further reinforce Aptamer C’s superior binding capability. Aptamer C forms 6 salt bridges with CD16 and 2 with PD-L1, surpassing all other aptamers. In comparison, Aptamer A forms only 3 salt bridges with each receptor, and Aptamer B forms 3 with CD16 and 5 with PD-L1, displaying an imbalanced binding profile. Aptamer D is the weakest candidate, forming just 2 salt bridges with CD16 and none with PD-L1, completely ruling it out as a viable bispecific aptamer. Since effective dual-targeting requires strong interactions with both receptors, Aptamer D’s inability to bind PD-L1 eliminates it from consideration.
Cation interactions and hydrophobic interactions were deemed irrelevant in determining the optimal bispecific aptamer due to their inconsistent presence across receptor interactions. Hydrophobic interactions were observed only with CD16, while cation-π interactions were exclusive to PD-L1, indicating that neither interaction type played a significant role in stabilizing dual-receptor binding. Hydrophobic interactions typically contribute to molecular stability in nonpolar environments, but their selective occurrence with CD16 suggests they do not meaningfully impact the overall binding affinity. Similarly, cation-π interactions, which involve electrostatic attraction between positively charged residues and aromatic rings, appeared only in PD-L1 interactions, making them unreliable as a universal stabilizing force. Given that hydrogen bonding and salt bridge formation were the dominant contributors to aptamer stability, hydrophobic and cation-π interactions were not considered essential in evaluating bispecific binding efficiency.
Electrostatic Surface Potential.
The ESP analysis and molecular docking results revealed key insights into aptamer binding efficiency to CD16 and PD-L1. Figure 4 shows that the binding regions of both receptors are predominantly positively charged, making them ideal for interaction with the negatively charged phosphate backbone of the aptamers. However, Aptamers B and D proved ineffective due to poor electrostatic complementarity and weak receptor interactions. Aptamer B’s misalignment with PD-L1 caused it to loop through the receptor twice, suggesting non-specific interactions that reduce accessibility to CD16, weakening bispecific binding. Additionally, this irregular binding pattern may lead to lower affinity and instability, making Aptamer B less reliable for therapeutic applications. Aptamer D also exhibited weak electrostatic attraction, resulting in no hydrogen bonding and salt bridge formation with PD-L1, further diminishing its effectiveness.
DISCUSSION.
While computational tools like AlphaFold 3, GrASP, and ChimeraX offer powerful insights into protein structure and potential interactions, they have limitations. Since this work is primarily computational, it relies heavily on algorithms and models that, while highly advanced, may not fully capture the complexities of biological systems (Kune, Haler, Far, & De Pauw, 2018). The accuracy of predictions, such as binding sites or ESP, depends on the quality of the data and the assumptions built into these models (Kune et al., 2018). In future studies, laboratory experiments are necessary to confirm the aptamer binding to the protein surface (Jhoti, Cleasby, Verdonk, & Williams, 2007). While computational methods offer a valuable starting point, real-world testing is crucial for confirming their findings.
Additionally, the receptor and aptamer binding orientation will make it difficult for both receptors to bind simultaneously. However, to computationally simulate bispecific binding, we would need to incorporate the plasma membrane on the surface-bound receptor and then dock them together. This approach is beyond the scope of our current research. In future studies, we plan to perform computational simulations that incorporate the plasma membrane using the CHARMM-GUI interface to analyze bispecific aptamer interactions more accurately. Future computational studies incorporating membrane-bound receptor models and flexible docking techniques will provide deeper insights into whether simultaneous binding to CD16 and PD-L1 is sterically and energetically feasible.
CONCLUSION.
This study demonstrates that bispecific aptamers targeting CD16 and PD-L1 can enhance NK cell-mediated cancer therapy. Among the aptamers analyzed, we found that Aptamer C exhibited the highest binding affinity and stability, suggesting its potential for clinical translation. While these findings are promising and open new doors for targeted cancer treatments, real-world experiments will be crucial to turning these concepts into practical therapies.
ACKNOWLEDGMENTS.
We sincerely thank Theodore Roosevelt High School, Des Moines, IA, for encouraging our research interest. We are especially grateful to Eigen Sciences LLC, Cary, NC, for their mentorship and guidance.
High-resolution aptamer images can be found here at our github: https://github.com/Babuji16/Aptamers”
REFERENCES.
- Mohanty, R., Chowdhury, C. R., Arega, S., Sen, P., Ganguly, P., & Ganguly, N. (2019). CAR T cell therapy: A new era for cancer treatment. Oncology Research, 42(6), 2183-2195.
- Miliotou, A. N., & Papadopoulou, L. C. (2018). CAR T-cell therapy: A new era in cancer immunotherapy. Current Protein & Peptide Science, 19(1), 5-18.
- Vormittag, P., Gunn, R., Ghorashian, S., & Veraitch, F. S. (2018). A guide to manufacturing CAR T cell therapies. Current Opinion in Biotechnology, 53, 164-181.
- Sterner, R. C., & Sterner, R. M. (2021). CAR-T cell therapy: Current limitations and potential strategies. Blood Cancer Journal, 11, 69.
- Anderson, J. K., & Mehta, A. (2019). A review of chimeric antigen receptor T-cells in lymphoma. Expert Review of Hematology, 12(7), 551-561.
- Guan, J., Lim, K. S., Mekhail, T., & Chang, C.-C. (2017). Programmed death ligand-1 (PD-L1) expression in the programmed death receptor-1 (PD-1)/PD-L1 blockade: A key player against various cancers. Annals of Pathology & Laboratory Medicine, 141(6), 851-861.
- Hudson, K., Cross, N., Jordan-Mahy, N., & Leyland, R. (2020). The extrinsic and intrinsic roles of PD-L1 and its receptor PD-1: Implications for immunotherapy treatment. Frontiers in Immunology, 11, 568931.
- Ortaldo, J. R., Mason, A. T., & O’Shea, A. (1995). Receptor-induced death in human natural killer cells: Involvement of CD16. The Journal of Experimental Medicine, 181(1), 339-344.
- Banerjee, J., & Nilsen-Hamilton, M. (2013). Aptamers: Multifunctional molecules for biomedical research. Journal of Molecular Medicine, 91, 1333-1342.
- Lee, J. F., Stovall, G. M., & Ellington, A. D. (2006). Aptamer therapeutics advance. Current Opinion in Chemical Biology, 10(3), 282-289.
- Song, K.-M., Lee, S., & Ban, C. (2012). Aptamers and their biological applications. Sensors, 12(1), 612-631.
- Rozenblum, G. T., Lopez, V. G., Vitullo, A. D., & Radrizzani, M. (2016). Aptamers: Current challenges and future prospects. Expert Opinion on Drug Discovery, 11(2), 127-135.
- Zheng, A., Du, Y., Wang, Y., Zheng, Y., Ning, Z., Wu, M., … & Liu, X. (2022). CD16/PD-L1 bi-specific aptamer for cancer immunotherapy through recruiting NK cells and acting as immune checkpoint blockade. Molecular Therapy – Nucleic Acids, 27, 998-1009.
- Fan, J., Fu, A., & Zhang, L. (2019). Progress in molecular docking. Quantitative Biology, 7, 83-89.
- Batta, I., & Sharma, G. (2024). Molecular docking simulation to predict inhibitors against zinc transporters. Computational Biology, 19, 100-112.
- Abramson, J., Adler, J., Dunger, J., Evans, R., Green, T., Pritzel, A., … & Bambrick, J. (2024). Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature, 630, 493-500.
- Xu, X., & Chen, S.-J. (2021). Predicting RNA scaffolds with a hybrid method of Vfold3D and VfoldLA. In RNA Scaffolds: Methods and Protocols (pp. 1-11). Springer.
- Yan, Y., Zhang, D., Zhou, P., Li, B., & Huang, S.-Y. (2017). HDOCK: A web server for protein–protein and protein–DNA/RNA docking based on a hybrid strategy. Nucleic Acids Research, 45(W1), W365-W373. doi:10.1093/nar/gkx407
- Yan, Y., Tao, H., He, J., & Huang, S.-Y. (2020). The HDOCK server for integrated protein–protein docking. Nature Protocols, 15, 1829-1852.
- Kune, C., Haler, J. R., Far, J., & De Pauw, E. (2018). Effectiveness and limitations of computational chemistry and mass spectrometry in the rational design of target‐specific shift reagents for ion mobility spectrometry. Chemistry, 19(21), 2921-2930.
21. Jhoti, H., Cleasby, A., Verdonk, M., & Williams, G. (2007). Fragment-based screening using X-ray crystallography and NMR spectroscopy. Current Opinion in Chemical Biology, 11(5), 485-493.
Posted by buchanle on Friday, June 20, 2025 in May 2025.
Tags: Aptamer, CAR T-cell Therapy, Molecular Docking, NK Cells

