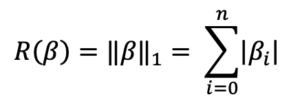Predicting Ovarian Cancer Using Regularized Logistic Regression
ABSTRACT
Ovarian cancer affects thousands of women annually. Currently, there is no screening test that is widely recommended for early detection of ovarian cancer. However, the analysis of certain biomarkers has demonstrated promising prospects in the prediction of ovarian cancer. Previous studies have selected various combinations of features in order to build predictive machine learning models using logistic regression or decision tree analysis. In an effort to select important features and to predict cases of ovarian cancer within a single unified framework, we proposed a logistic regression model with a LASSO regularization penalty. The resulting model selected 30 features with significance in predicting ovarian cancer (including three clinically relevant biomarkers: HE4, CA125, and CEA) and demonstrated high accuracy, sensitivity, and specificity. The results demonstrate the viability of using a logistic regression model with a LASSO penalty and provide a baseline for further research involving cross-validation.
INTRODUCTION.
Ovarian cancer has the fifth highest mortality rate of all cancers among women. The National Cancer Institute predicted that in 2021, there would be a total of 21,410 cases and 13,770 deaths attributed to ovarian cancer [1]. Unfortunately, there are currently no screening tests that professionals have widely recommended to detect ovarian cancer in its early stages [2]. It is critical to improve methods for the early detection of ovarian cancer, as accurate screening has been projected to reduce mortality by 10% to 30% [3]. Many groups have investigated biomarkers to predict ovarian cancer, which is proving to be a promising area for further research [4-8].
Many screening strategies have involved testing patients’ blood for the presence of biomarker protein cancer antigen 125 (CA125), which is often produced in higher levels in women with ovarian cancer and has proven to serve as a robust predictor for ovarian cancer [8, 9]. However, better prediction accuracy, particularly in earlier stages, has been achieved when evaluation of CA125 is used in conjunction with other strategies such as transvaginal sonography or other biomarkers such as human epididymis protein 4 (HE4), which is produced by many epithelial ovarian cancer cells [4, 8].
In one key study on predicting ovarian cancer, Lu et al. chose to evaluate 49 features ranging from biomarkers to patient characteristics such as age or blood routine test results to develop their model [10]. They implemented a form of filter type feature selection in order to determine which of these features were most significant to aid with their prediction and used a decision tree to predict an individual’s likelihood of developing ovarian cancer under the final subset of features [10]. More specifically, they used a combination of Minimum Redundancy Maximum Relevance (MRMR) feature selection, ReliefF feature selection, and decision tree analysis to carry out their modeling procedure [10]. Ultimately, they found that a decision tree approach using HE4 and carcinoembryonic antigen (CEA) could be used to achieve a predictive accuracy of 92.1% [10].
In an effort to simultaneously select important features and predict risk of ovarian cancer within a single unified framework, we propose constructing a logistic regression model with a Least Absolute Shrinkage and Selection Operator (LASSO) regularization penalty. LASSO is a regularization penalty that operates on the objective function of well-studied regression problems. Under LASSO, models such as logistic regression can estimate their parameters while simultaneously selecting features that are important (see Methods). This is a critical distinction from the Lu et al. model as we avoid treating feature selection and model prediction as two separate processes, instead consolidating them under a single, unified model. Using this approach, we aim to achieve a predictive performance comparable to that of the Lu et al. model while also selecting a set of significant features that encompasses those originally selected. The ultimate intent of our work is to predict whether or not an individual is likely to eventually be diagnosed with ovarian cancer based on their biological data.
MATERIALS AND METHODS.
Data.
The dataset used in this study consists of data sampled from 349 patients (171 patients with ovarian cancer and 178 patients with benign ovarian tumors) from the Third Affiliated Hospital of Soochow University, enrolled between July 2011 and July 2018 [10]. Following the patients’ surgical resection, diagnostic pathology identified each patient as belonging to one of two groups: either Benign Ovarian Tumor (BOT) or Ovarian Cancer (OC) [10]. Biological information collected from each patient included blood routine tests, general chemistry tests, and tumor marker analysis [10]. The dataset includes 49 potentially relevant features such as demographic, age, blood routine test results, and various biomarkers [10]. Lu et al. sought to use this dataset in order to perform filter type feature selection and develop a model for predicting ovarian cancer [10].
To prepare the data, we performed the following data pre-processing steps: First, we removed data points that contained inappropriately labeled information. We also removed any columns that contained a significant amount of missing data, namely biomarker columns NEU and CA72-4, and imputed any remaining missing information using the mean values computed from the training data. Furthermore, we standardized our features to have zero mean and unit variance, a requirement for the regularization penalty discussed in our Methods section.
In their study, Lu et al. partitioned their dataset into training and testing sets. We combined these datasets into a single set and randomly selected 70% of the data points to make up the training data. The remaining values were used for testing. We further divided the training data according to an 80:20 ratio to allow for hyperparameter tuning. After this further partitioning, 80% of the data (191 data points) made up the sub-training set, and 20% (48 data points) made up the validation set.
Methods.
In order to achieve more accurate predictions with our logistic regression model, we applied regularization with an L1 penalty. Generally, regularization is the process by which a model’s coefficients, or weights, are penalized [11]. This approach prevents the model from being overfitted to the training data, which could result in greater inaccuracy when we apply the model to previously unseen data [11]. When λ, the LASSO tuning parameter, is zero, the model performs multiple logistic regression with no penalization. When λ is very large, the absolute values of the coefficients all shrink to zero [11]. This means that LASSO regression also serves as an effective form of feature selection because, for an appropriate choice of λ, irrelevant features immediately shrink to zero (deselection), whereas the significant features remain non-zero (selection). To ensure the LASSO penalty is evenly applied to each coefficient, we must standardize our features. The LASSO penalty utilizes the L1 norm as follows, where \[\beta\] is a regression coefficient:
To determine the most appropriate regularization strength, we iterated through multiple values of λ and evaluated the predictive accuracy on the validation data for each one.
RESULTS.
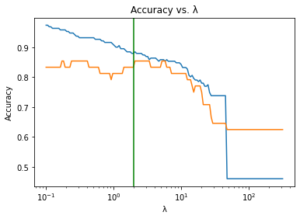
In order to select the appropriate value of λ to achieve a model with the highest possible predictive accuracy, we divided the preprocessed data into training and testing datasets. We further divided the training data into sub-training and validation datasets. We iterated through various values of λ using both the sub-training and validation datasets and identified λ = 2 as the most appropriate regularization strength (see Figure 1).
Figure 1. KEY – Blue: sub-training accuracy, Orange: validation accuracy, Green: λ = 2; A graph generated by iterating through various values of λ (tuning parameter) and finding their corresponding accuracies in predicting ovarian cancer diagnosis. λ = 2 was determined to be the most desirable because it maximizes both validation and sub-training accuracy while maintaining minimal difference between them.
Our goal is to maximize accuracy for both the validation and sub-training data while maintaining minimal difference between them. When λ equals 2, both of these criteria are true, indicating that the highest predictive accuracy should occur around this point. When we fitted a logistic regression model to the testing data using a LASSO penalty of 2, our model obtained an accuracy of 90.6%. The model selected 30 features, including HE4, CA125, and CEA.
We calculated a few key diagnostics to evaluate the success of the model. As stated, its accuracy was 90.6%, indicating that it was able to predict the correct outcome 90.6% of the time. Its sensitivity was 86.8%, indicating that it correctly predicted 86.8% of all ovarian cancer outcomes. Finally, its specificity was 100.0%, indicating that it correctly predicted all benign tumor outcomes. All three metrics demonstrate high values.
DISCUSSION.
Lu et al. implemented a Minimum Redundancy-Maximum Relevance feature selection that sought to ensure that the selected features were mutually maximally dissimilar. However, their approach involved a complicated process of dividing the available data into ten sample groups (significantly reducing the already-small sample size), selecting features from each sample group, and then constructing a 5-level decision tree model. They then repeated this process ten times to find the highest accuracy. Ultimately, Lu et al. determined that they were able to achieve an accuracy of 94.3% on their validation data when the model utilized the selected features CEA and HE4.
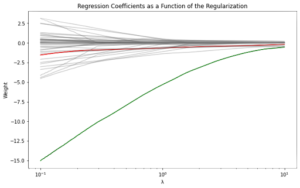
The approach taken in our study is distinctive from that of Lu et al.’s due to the simplistic nature of the logistic regression model with a LASSO penalty. Our model was developed without requiring division into even smaller samples or the construction of a complex decision tree. We were also able to achieve a predictive accuracy on our validation data of 90.6%, which is relatively comparable to the accuracy achieved by Lu et al. Furthermore, our study’s feature selection, while simpler than Lu et al.’s, did successfully select both CEA and HE4 in addition to other biomarkers selected in previous studies, such as CA125 (see Figure 2).
Figure 2. KEY – Red: coefficient magnitude of biomarker CEA, Green: coefficient magnitude of biomarker HE4; A demonstration of how regression coefficients shrink to zero as λ, the regression model tuning parameter, increases. This enables LASSO to select features because, at an appropriate choice of λ, significant features will remain non-zero. In the graph, HE4’s high magnitude indicates its significance. CEA has been selected because it is non-zero, but it is not particularly significant.
Regularizing logistic regression has the ability to shrink coefficients with respect to λ. In examining this trend, we observe that some of the features selected in previous studies were resistant to shrinking to zero for higher values of λ, highlighting the significance of these particular features in predicting ovarian cancer.
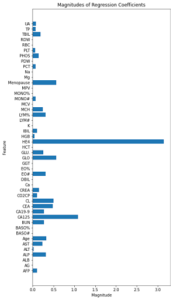
In order to further investigate the significance of certain features, we evaluated the magnitudes of their regression coefficients (see Figure 3).
Figure 3. Absolute magnitudes of the regression coefficients. Higher magnitude indicates greater influence on predictive outcomes, while a magnitude of zero signifies a deselected feature.
Through our literature review, we found that HE4 is a biomarker that is commonly identified by previous studies to have high significance in predicting ovarian cancer. In accordance with this finding, HE4 has a significantly high coefficient magnitude compared to all other features. CA125, another feature commonly utilized in previous studies, has the second highest coefficient magnitude. CEA, the other feature selected by Lu et al. in addition to HE4, does not necessarily demonstrate a high coefficient magnitude when compared to the other features. However, the observable magnitude indicates that the model identified it as having some significance in ovarian cancer prediction.
Furthermore, the high values for the key model diagnostics—accuracy, sensitivity, and specificity—suggest that the model offers a novel method of accurately predicting ovarian cancer outcomes with low risk for false positives or false negatives. If either sensitivity or specificity were significantly low, it would have limited the model’s usefulness in certain situations. However, these metrics indicate that the model may be useful in a variety of contexts, as a high degree of trust can be placed in the predicted outcome, whether it is a benign ovarian tumor or ovarian cancer.
The results obtained through our model demonstrate the viability of using a logistic regression model with a LASSO penalty in order to predict an individual’s likelihood of developing ovarian cancer based on various features of their biological data.
Limitations.
The main limitation encountered in this study was the small size of the original dataset, which consequently made the training and testing sample sizes small as well. Due to this limitation, we found that splitting the data in different ways, namely using different seeds for the random sampling of the training dataset, resulted in different predictive accuracies. The small number of data points available for model training resulted in consistent overfitting. Ideally, we would have liked to perform the study using a larger dataset had one been available.
Future Approaches.
In the future, it may be worthwhile to repeat this study with the incorporation of cross-validation, which may help to mitigate the consequences of using a small dataset and prevent the developed model from overfitting the training data. In five-fold cross-validation, for example, we would split the data into five subsets, treating four of them as training data and the fifth as testing data. We would fit a model using these parameters, retaining the resulting evaluation score. After repeating this process five times, each time with a different test subset, we would ultimately determine the model’s accuracy based on the five evaluation scores. If this approach successfully prevents overfitting, it could serve as a valuable method to improve this study’s findings by identifying a model that is more widely applicable to previously unseen datasets.
CONCLUSION.
Inspired by the research performed by Lu et al. in their paper, “Using machine learning to predict ovarian cancer,” we sought to achieve a similar aim, namely the prediction of ovarian cancer, using a simpler, more convenient method of feature selection and model development. Using a logistic regression model with a LASSO penalty, we were able to achieve a predictive accuracy of 90.6% while also selecting the same features selected by Lu et al. in their study. Our findings demonstrate the potential for LASSO logistic regression in developing a machine learning model to predict an individual’s likelihood of future ovarian cancer.
ACKNOWLEDGMENTS.
I thank Mr. Patrick Emedom-Nnamdi from the Harvard University Department of Biostatistics for his guidance throughout my research. I also thank Mr. David Hansen for editing this paper and my family for their constant support.
GLOSSARY.
Accuracy: Ratio of the number of correct results to the total number of results
L1 Penalty: Sum of the coefficient magnitudes in a LASSO regression model; facilitates regularization
λ (Lambda): Tuning parameter in a LASSO regression model whose increase causes coefficient magnitudes to shrink towards zero; facilitates feature selection
LASSO: Regularization method that incorporates an L1 penalty to perform feature selection with respect to a tuning parameter, λ
Logistic Regression: Predictive statistical method that models the conditional probability of a binary response variable Y given a value of X
Regularization: Process by which a model’s coefficients are penalized; prevents overfitting to training data
Sensitivity: Measure of true positives; ratio of the number of those predicted to have target disease to the total number of those with target disease
Specificity: Measure of true negatives; ratio of the number of those predicted to not have target disease to the total number of those without target disease
Training Dataset: Example data to which a machine learning model is fitted
Validation Dataset: Example data on which a fitted machine learning model is tested in order to provide an estimate of prediction error
REFERENCES.
- American Cancer Society PDQ Adult Treatment Editorial Board, “Ovarian Epithelial, Fallopian Tube, and Primary Peritoneal Cancer Treatment”.
- American Cancer Society Medical and Editorial Content Team, “Cancer Facts & Figures 2021”.
- D. A. Fishman, K. Bozorgi, “The Scientific Basis of Early Detection of Epithelial Ovarian Cancer: The National Ovarian Cancer Early Detection Program”. Cancer Treat. Res. 107, 3–28 (2002).
- R. C. Bast, Z. Lu, C. Y. Han, K. H. Lu, K. S. Anderson, C. W. Drescher, S. J. Skates, “Biomarkers and strategies for early detection of ovarian cancer”. Cancer Epidemiol., Biomarkers Prev. 29, 2504–2512 (2020).
- N. Banaei, J. Moshfegh, A. Mohseni-Kabir, J. M. Houghton, Y. Sun, B. Kim, “Machine learning algorithms enhance the specificity of cancer biomarker detection using SERS-based immunoassays in microfluidic chips”. RSC Adv. 9, 1859–1868 (2019).
- J. Ma, J. Yang, Y. Jin, S. Cheng, S. Huang, N. Zhang, Y. Wang, “Artificial Intelligence Based on Blood Biomarkers Including CTCs Predicts Outcomes in Epithelial Ovarian Cancer: A Prospective Study”. OncoTargets Ther. 14 (2021).
- P. N. Yeganeh, M. T. Mostafavi, “Use of Machine Learning for Diagnosis of Cancer in Ovarian Tissues with a Selected mRNA Panel”. Proc. – 2018 IEEE Int. Conf. Bioinf. Biomed. 2018, 2429–2434 (2019).
- R. G. Moore et al., “Comparison of a novel multiple marker assay vs the Risk of Malignancy Index for the prediction of epithelial ovarian cancer in patients with a pelvic mass”. Am. J. Obstet. Gynecol. 203, 228.e1-228.e6 (2010).
- H. J. Whitwell, et al., “Improved early detection of ovarian cancer using longitudinal multimarker models”. Br. J. Cancer. 122, 847–856 (2020).
- M. Lu, et al., “Using machine learning to predict ovarian cancer”. Int. J. Med. Inf. 141 (2020).
- G. James, D. Witten, T. Hastie, R. Tibshirani, An Introduction to Statistical Learning. (Springer New York, New York, NY, 2013).
- R. Chen, “Machine learning for ovarian cancer: lasso regression-based predictive model of early mortality in patients with stage I and stage II ovarian cancer”. medRxiv. (2020).
- American Cancer Society Medical and Editorial Content Team, “About Ovarian Cancer”.
- S. W. Lee, H. Y. Lee, H. J. Bang, H. J. Song, S. W. Kong, Y. M. Kim, “Improved Prediction Model for Ovarian Cancer Using Urinary Biomarkers and a Novel Validation Strategy”. Int. J. Mol. Sci. 20 (2019).
- H. Zhao, H. Hayat, X. Ma, D. Fan, P. Wang, A. Moore, “Molecular imaging and deep learning analysis of uMUC1 expression in response to chemotherapy in an orthotopic model of ovarian cancer”. Sci. Rep. 10, 1–13 (2020).
- M. Wu, C. Yan, H. Liu, Q. Liu, “Automatic classification of ovarian cancer types from cytological images using deep convolutional neural networks”. Biosci. Rep. 38 (2018).
- K. M. Elias et al., “Diagnostic potential for a serum miRNA neural network for detection of ovarian cancer”. eLife. 6 (2017).
- L. Y. Guo, A. H. Wu, Y. X. Wang, L. P. Zhang, H. Chai, X. F. Liang, “Deep learning-based ovarian cancer subtypes identification using multi-omics data”. BioData Min. 13, 1–12 (2020).
- M. Akazawa, K. Hashimoto, “Artificial Intelligence in Ovarian Cancer Diagnosis”. Anticancer Res. 40, 4795–4800 (2020).
- A. Laios et al., “Feature Selection is Critical for 2-Year Prognosis in Advanced Stage High Grade Serous Ovarian Cancer by Using Machine Learning”. Cancer Control. 28 (2021).
- R. M. Ghoniem, A. D. Algarni, B. Refky, A. A. Ewees, “Multi-Modal Evolutionary Deep Learning Model for Ovarian Cancer Diagnosis”. Symmetry. 13, 643 (2021).
- K. H. Yu, V. Hu, F. Wang, U. A. Matulonis, G. L. Mutter, J. A. Golden, I. S. Kohane, “Deciphering serous ovarian carcinoma histopathology and platinum response by convolutional neural networks”. BMC Med. 18, 1–14 (2020).
- P. Zhang, et al., “Development of a multi-marker model combining HE4, CA125, progesterone, and estradiol for distinguishing benign from malignant pelvic masses in postmenopausal women”. Tumour Biol. 37, 2183–2191 (2016).
Posted by John Lee on Saturday, May 7, 2022 in May 2022.
Tags: LASSO, Logistic regression, Ovarian cancer