Isolating Endophytes to Derive Novel Antibiotics
ABSTRACT
With about 33,000 deaths every year from infections caused by resistant bacteria, the need for new antibiotics is growing. In order to find new antibiotics, this study looked to bacteria and fungi, specifically those found in medicinal plants, because most current antibiotics were derived from bacteria and fungi. The microorganisms studied in this paper were extracted from the plants Mentha spicata, Mentha piperita, Rosmarinus officinalis, and Ginkgo biloba because these plants are known to have antibiotic properties. In total, 66 bacterial samples and 86 fungal samples were collected. Furthermore, the majority of these samples belonged to intercropped plants (i.e. plants grown among other plant species). Of the bacteria, thirtyamples were tested for resistance to current antibiotics. This test was to identify microorganisms with strong defense mechanisms. In this test it was found that four of these samples had some form of resistance to either penicillin, novobiocin, chloramphenicol, or erythromycin. Also, these thirty samples along with four fungal samples were tested for their interactions with Escherichia coli. This test found six samples (four bacteria and two fungi) that resisted the bacteria. These resistant strains are significant because they show to have a defense mechanism against possible antibiosis. If the defense mechanism is a chemical secretion, then the future isolation of this chemical compound will lead to the advent of a novel antibiotic.
INTRODUCTION.
In an age where many of the greatest health problems were either eradicated or are being eradicated, approximately 33,000 people die every year from infections caused by antibiotic resistant bacteria [1]. To fix this problem, we need new antibiotics,but drug companies and manufacturers are not producing novel products because the cost of development is too high [2].
All of our current antibiotics were derived from naturally produced compounds [3]. These compounds were originally defense mechanisms for their source microorganisms. For example, the common antibiotic penicillin is manufactured from a compound naturally produced by the penicillium mold. This compound acts as a defensive mechanism against surrounding bacteria. Likewise, this study aimed to identify microorganisms that synthesize bacteria resistant compounds.
In order to find these microorganisms, we decided to look in plants as plants and bacteria/fungi often form mutualistic relationships. Furthermore, we looked in medicinal plants because they are known to contain easily isolatable endophytes (microorganisms that live within plants) and are more likely to contain endophytes with antibiotic properties [4]. The four species were selected according to availability and known antibiotic properties: Mentha spicata, Mentha piperita, Rosmarinus officinalis, and Ginkgo biloba [5-7]. All four of these plants have been used in traditional medicine for centuries and thus were the best place to begin looking for these necessary microorganisms.
The two tests used to determine whether or not each endophyte produced a bacteria resistant compound were an antibiotic susceptibility test and a test against Escherichia coli. First, antibiotics work similarly to defensive bacterial compounds in that their mechanisms’ goal is to reduce surrounding bacteria. A bacteria resistant to this compound has a defense mechanism that defeats the antibiotic’s compound. This defense mechanism may be a compound that itself is antibiotic. Second, a test against another bacteria, E. coli, clearly demonstrates the strength of the bacteria’s— the one being tested—antibiotic compound.
In addition, this study examined the environmental impact on bacterial and fungal diversity of endophytic microbiomes. By collecting microorganisms from both environments that were monocropped (only one plant) and intercropped (a plant cultivated with others), we aimed to determine which of the two environments produced more diversity. We hypothesized that the intercropped samples would be more diverse because they have a rich phytobiome influence [8].
MATERIALS AND METHODS.
Isolating Single Endophytic Species.
One empty Petri dish was used to rinse and surface sterilize each piece of the plant samples. For the R. officinalis, M. spicata, and M. piperita only the stem and leaf were surface sterilized. For the G. biloba, the wooded stem, leaf, and green stem were prepared. For sterilization, the samples were soaked in 75% ethanol and then rinsed three in reverse osmosis (RO) treated sterilized water for 1 minute. Each part of the four plants were sliced into the following: (1) 1 mm segments for the stems (2) 1 mm segments for the wooded G. biloba stem after being debarked (3) about 1 mm by 1 mm squares for the leaves. Each part of each plant was placed in both a Sabouraud dextrose agar plate and a Wallerstein nutrient agar plate in order to ensure the growth of both bacteria and fungi. Each plate was sealed with parafilm to prevent contamination, then incubated at room temperature (28°C).
For the next two weeks, plates were monitored for contamination and, when appropriate, subcultured. Subculturing occurred when at least two distinct microbial species were visible in one Petri dish. Subculturing was performed until definite, pure isolates of either fungi or bacteria are achieved on their respective dishes.
Characterizing the Bacteria.
A single colony was used to carry out a gram stain reaction. The gram stain tests were performed on 30 bacterial samples. The bacteria were selected based on which plant they were collected from and by the part of the plant. The morphological characteristics of these isolates was recorded as well.
The 30 bacterial samples were tested for resistance to four antibiotics: chloramphenicol (15μg), erythromycin (15μg), penicillin (10μg), novobiocin (30μg). The results were recorded.
Testing Bacterial and Fungal Isolates against E. coli.
A single E. coli colony was grown in a shaker to achieve McFarland standard, then spread on a Müeller-Hinton (MH) agar plate to create a lawn. Fifteen of the thirty bacterial samples were also grown in 50mL Luria Broth keeping in mind the representation of each plant and plant part. Blank sterile disks were impregnated with 25µl of each of the fifteen bacteria in triplicates. The disks were then planted onto the MH agar plates with E.coli in triplicates for each bacterial isolate. The plates were left to grow at 25 °C for approx. 48 hours. The zones of inhibition were measured and recorded according to CLSI (Clinical and Laboratory Standards Institute) standards [9]. Resistance was determined based on the sizes of the zones of inhibition.
A single E. coli colony was prepared to McFarland standard and plated onto a MH agar plate to prepare a lawn. This was done in four replicates. Using a cork borer, three disks were punched per fungus and placed on the lawn of E. coli on the MH plate. This was done in triplicates. The plates were left to grow at 25 °C for 48 hours. Resistance was measured by observing the zones of inhibition according to CLSI standards.
RESULTS.
This section provides data for the main hypothesis of this study: intercropped environments will produce more diverse microorganims than monocropped environments. Also, information characterizing the collected microorganisms is presented here.
Endophytes isolated from Monocropped versus Intercropped Plant Environments.
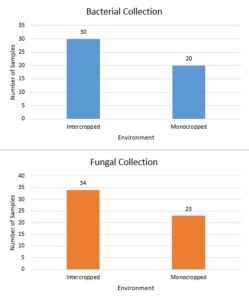
Figure 1 shows that 57 fungal and 50 bacterial endophytes were extractedfrom plants were both mono and intercropped samples were available. Only in M. spicata did the intercropped samples produce more isolates of fungi and bacteria than monocropped samples. Monocroppred M. piperita and R. officinalis produced more isolates than their intercrop counterparts. While the pool data suggests that intercropping produces a more diverse endophytic community, the per plant data suggests that that conclusion would vary plant to plant.
Figure 1. Total endophytes isolated based on plant environment from R. officinalis, M. spicata, and M. piperita.
No monocropped samples of G. biloba were used due to unavailability. As such, samples collected from G. biloba were excluded from the data.
Gram Stain and Morphology.
Gram staining and morphology analysis of 30 bacterial samples isolated from various parts of all 4 plants species collected foundmost of the bacteria are Gram-positive bacilli, as shown in Table 1. The data shows a diverse set of bacteria.
Table 1. Results of the Gram stain tests for each bacteria tested.
| Gram and Shape | M. Spicata | M. piperita | R. officinalis |
G. biloba
|
| Bacilli + | 4 | 12 | 1 | 5 |
| Bacilli – | 0 | 1 | 0 | 1 |
| Cocci + | 0 | 0 | 0 | 0 |
| Cocci – | 0 | 5 | 0 | 1 |
Antibiotic Resistance Tests.
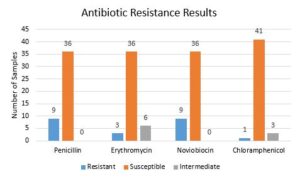
The antibiotic test results showed that some bacterial isolates were resistant to common antibiotics. Several of the bacteria were resistant or intermediate on more than one antibiotic as shown in Figure 2. This study is primarily interested in microorganisms that resistant these antibiotics as those resisting are likely have a defense mechanism that produces a resistant compound. As a result, this test is a first step in identifying microorganism that produce antibiotic materials.
Figure 2. The results of the antibiotic resistance tests.
Resistance to E. coli.
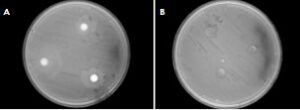
Only four bacteria and two fungi tested against E. coli were found to create zones of inhibition. The best sample of bacteria (A) and the best of the fungi (B) are shown in Figure 3. This data shows the best six specimens for further experimentation.
DISCUSSION.
As Figure 1 shows, the pool of data demonstrates that more endophyte isolates were collected from intercropped plants. This result was predicted as growing in a more diverse environment allowed the plants to be exposed to more microorganisms. However, in M. spicata more fungal and bacterial endophytes were isolated from intercropped plants versus monocropped plants. While this result does not negate the conclusion made previously, it suggests that there may be factors other than plant diversity that determine the microbial diversity of a system. One possibility is that the increased diversity of the soil makes it more likely that a single dominant species become prominent in that patch of soil. Thus, this test will need to be repeated in order to account for the location’s effect on the number of bacterial and fungal isolates.
The results displayed in Table 1 describe some characteristics of the bacteria collected which will allow for the identification of these bacteria in future studies.
The results of the antibiotic resistance tests shown in Figure 2 are mixed. Most bacteria were susceptible to all four antibiotics used. The few that were resistant to any of the antibiotics need to be identified with a DNA test so that (1) individuals in the medical field are aware that these bacteria strains are not affected by certain antibiotics and (2) future studies may try to identify the defense mechanism. These samples may lead to the creation of new antibiotics if it is discovered that their defense mechanisms are antibiotic chemical secretions.
The interaction tests, as displayed in Figure 3, showed very few successful trials—that is, trials in which the bacteria/fungal isolate created a zone of inhibition. Trials that produced these zones are to be further tested to confirm which entity, the isolate being tested or the E. coli, is producing the inhibiting agent. Finding which species is the causing agent is crucial because the lawn of E. coli and the bacteria being tested were grown at the same time. Because of this, either could be inhibiting the other. If the isolate being tested is producing the inhibiting agent, then it is killing the E. coli. If the E. coli is producing the inhibiting agent, then the effect being measured is the E. coli repelling our isolate. The procedure also needs to be repeated to test against other species of illness causing bacteria.
Figure 3. The most resistant bacteria (A) and fungus (B) to E.coli with zones of inhibition surrounding the disks containing tested endophytes.
CONCLUSION.
The primary continuation of this work is the DNA extraction and sequencing of each isolate in order to identify the endophytic species for phylogenetic analysis. Also, the exploration of seasonal changes and their effects on endophytic diversity, the isolation of alkaliphiles (bacteria that grow in basic conditions), and sampling the same plant species from different locations will provide data on both endophytic diversity and new endophytic samples. Finally, further in vitro interaction of the endophytes from the same microbiome and the interaction between the endophytic isolates with other clinically-relevant microorganisms will help to determine other valuable candidates for antibiotic synthesis.
The most important of these future directions is the extraction and sequencing of DNA because that process will identify the fungi and bacteria found during this project to species level.
In conclusion, the collected data states in sum that endophytic diversity tends to be greater in plants that live amongst other plants, but this conclusion is not consistent in all cases . The exposure that they have to these phytobiome allows more fungi and bacteria to interact. Outside of this finding, more work needs to be done to conclude on the other topics of interest.
ACKNOWLEDGMENTS.
We would like to give special thanks to the following people and organizations for their help, support, and/or funding of this project: Belmont University SURF, Montgomery Bell Academy, the Wilson family, Nicholas Ragsdale PhD, Matthew Heard PhD, Rabab K. Sager PhD, Jonathan Creamer PhD, Jim Dickens PhD, Afaf Shamma MSc, and Luma S. Hachim MSc.
REFERENCES
- R. Leistner, Faculty of 1000 evaluation for Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. F1000 – Post-publication Peer Review of the Biomedical Literature (2018).
- Robert Langreth, Antibiotics Aren’t Profitable for Big Pharma to Make More, Bloomberg Businessweek (2019).
- A. L. Demain, S. Sanchez, Microbial Drug Discovery: 80 Years of Progress. J. Antibiot (Tokyo), 62, 5-16 (2009).
- M. Jia, L. Chen, H. Xin, C. Zheng, K. Rahman, T. Han, L. Qin, A Friendly Relationship between Endophytic Fungi and Medicinal Plants: A Systematic Review. Frontiers in Microbiology, 7, 906 (2016).
- R. Singh, M. A. Shushni, A. Belkheir, Antibacterial and antioxidant activities of Mentha piperita, L. Arabian Journal of Chemistry, 8, 322-328 (2015).
- Q. Liu, X. Meng, Y. Li, C. Zhao, G. Tang, H. Li, Antibacterial and Antifungal Activities of Spices. International Journal of Molecular Sciences, 18, 1283 (2017).
- Y. Wu, T. Zhang, M. Zhang, J. Cheng, Y. Zhang, An endophytic Fungi of Ginkgo biloba L. produces antimicrobial metabolites as potential inhibitors of FtsZ of Staphylococcus aureus. Fitoterapia, 128, 265-271 (2018).
- J. E. Leach, L. R. Triplett, C. T. Argueso, P. Trivedi, Communication in the Phytobiome, Cell Press, 587-596 (2017).
- Vitek, Antimicrobial Susceptibility Testing. Biomedical Safety & Standards, 49, 28 (2019).
Posted by John Lee on Wednesday, December 23, 2020 in May 2020.
Tags: Antibiotics, Endophytes, Medicinal Plants, Resistance