Effects of artificial algal-bacterial symbioses (E. Coli K-12, C. Fruendii, & E. Coli MC4100) on microbial hydrogen production
ABSTRACT
Climate change is a very current and real problem, and the most effective way to combat it would be to switch energy production from unsustainable fossil fuels to sustainable, renewable energy sources. One promising opportunity is hydrogen, as it burns clean and has high energy per volume. However, producing hydrogen gas is currently done via steam methane reformation which cuts into its benefits because it produces CO2.In order to form a hydrogen economy, a clean way of producing hydrogen must be developed. Various microbes produce hydrogen from their biological processes, and these processes, along with inter-microbial dynamics and relationships like symbiosis, can be exploited for human benefit. The purpose of this experiment is to research the potential of algal-bacterial symbiotic cultures on increasing hydrogen production and compare that to either organism individually. It is hypothesized that if hydrogenases are inhibited by higher oxygen levels and aerobic bacteria consume oxygen, then the symbiotic cultures will produce increased levels of hydrogen. Both pure algal and pure bacterial cultures were maintained and then combined and their H2 production analyzed via titration. With a p value overall of less than 0.05, the results are significant, and show that each of the presented bacterial species and strains work with Chlamydomonas algae for enhancing microbial hydrogen production. This information could be expanded upon and used to optimize microbial hydrogen production to the commercial scale, a step this field needs to take.
INTRODUCTION.
Hydrogen is being explored in energy as both a fuel itself and as a storage method because it is almost completely clean, that is, it releases little to no greenhouse gasses into the atmosphere upon combustion or other usage. It is also higher in energy than fossil fuels by mass. The main problem with hydrogen as a fuel is in regard to infrastructure and transportation; hydrogen has to be stored, and thus transported, at extremely low temperatures and specific pressures to be of any use, which current infrastructure is not built for[1]. By far its most common current use in energy is in fuel cells to power cars, which work through a positive anode and negative cathode being fed hydrogen and oxygen separately to release electrons that perform work, and then all combine to form water byproduct [2]. Pure hydrogen must be produced, which is almost always done through industrial means like steam reforming natural gas. This works by combining extremely hot steam, methane, and a catalyst, separating them, releasing carbon dioxide and other impurities, which leaves only pure hydrogen and contributes to the greenhouse gas effect [2].
Another method of producing algae comes from a potentially surprising source: microorganisms. Algae are a diverse group of photosynthetic eukaryotes found everywhere from backyard pools to open oceans. Bacteria are an incredibly old and diverse group of prokaryotes, with a unique subset, known as cyanobacteria, that undergo photosynthesis and consume oxygen. Together, algae and cyanobacteria are called phytoplankton and make up the basis of many ecosystems on earth, especially in oceans where they are the undisputed primary producers. Eukaryotic algae likely evolved from bacteria, and thus, have likely had symbiotic relationships for hundreds of millions of years. However, these relationships have only been the subject of research quite recently, both because of lack of insight to their importance and difficulty in conducting separate research on one organism since they rely so heavily on one another. Between the three main types of symbiotic relationships, mutualism, parasitism, and commensalism, parasitism is the most well studied and commensalism the least. Red algae are considered readily parasitic. Commensal relationships are hard to pin down and study, and mutualistic relationships typically revolve around some sort of substance exchange [3]. Algal-bacterial symbiosis always happens in close contact, so substances can readily transfer between organisms, whether as wastes or as by-products. These substances are typically vitamins produced by the bacteria, B vitamin cofactors especially, and are used by the algae, as well as dissolved nutrients (C, N, P, S, Fe), and consumed by the bacteria. This exchange fosters growth in both organisms [4]. “Artificial” symbiosis, or symbiosis not typically found in nature, has yet to be thoroughly studied, particularly in the field of hydrogen production [5].
Recall that the current most efficient and widely used method of producing hydrogen is industrial and an intensive process. Without a clean way to produce hydrogen, its benefits as a clean energy source are at best undermined and at worst nullified. Various organisms naturally produce hydrogen due to their internal biological processes, and these processes can be exploited and maximized in various ways for human benefit. The organisms best suited for biological hydrogen production are green algae, Cyanobacteria, photosynthetic bacteria, and dark fermentative bacteria, most notably because of their minimal nutritional requirements and potential hydrogenases [6].
Hydrogenases are enzymes or enzyme complexes that catalyze the simple, reversible reaction of the oxidation of H2 into two protons and two electrons, and as such, represent the star of the show on the stage of biological hydrogen production. FeFe hydrogenases are studied because they are the ones found in both bacteria and eukarya. (FeFe) hydrogenases have about a 100-fold higher activity than others, but are sensitive to oxygen; that is, if oxygen levels get high enough, the hydrogenase enzymes will be irreversibly inhibited, making them hard to use for H2 production under natural oxygen conditions. The most common way to counteract this is via sulfur deprivation, where depriving the culture of sulfur leads to total anoxia and the oxygen-sensitive FeFe hydrogenases become functional. This method can be cumbersome as it requires prior cultivation, change of media, which only then initiates the H2 generation stage [7]. One way to counteract the oxygen sensitivity of FeFe hydrogenase is to pair the organism with an aerobic one. That way, oxygen gets consumed and the overall level never gets high enough to inhibit the hydrogenases. This can be achieved by either natural or artificial symbiosis, which is the principle behind one study done at the University of West Hungary. The experimental setup consisted of two strains of Chlamydomonas algae cultured with either naturally symbiotic bacteria, artificially symbiotic bacteria (E. coli), or no bacteria (pure algae) at varying volumes and lengths of time. Consistently, the mixed cultures with E. Coli produced more hydrogen than the pure ones [5].
Escheria coli MC4100 is a particular strain of E.coli that has a different genome than the typical K-12 strain typically used experimentally. The difference relevant to this project is the lack of a functioning hypF gene, which is involved in the maturation and function of NiFe hydrogenases. While these aren’t FeFe hydrogenases, meaning they are not as efficient or as sensitive to oxygen, they do contribute to hydrogen production. Without this gene, these hydrogenases won’t function [8]. Including them in this experiment will provide insight into whether it is beneficial to have bacteria with functioning hydrogenases in tandem with algae, a pair greater than the sum of its parts, or whether it would just get in the way of Chlamydomonas hydrogenase up taking the finite reactants and nutrients.
Citrobacter freundii is a common member of the soil microbiome, playing an important role in the environmental nitrogen cycle through reduction of nitrate to nitrite [9]. Both Escherichia coli and Citrobacter freundii are facultative aerobic bacteria that are well understood and relatively simple to work with in the lab. In culturing these bacteria with the same kind of algae and seeing the differences in the effects, it can be better understood what exactly makes the difference when trying to enhance algal hydrogen production. The overlaps and differences can paint a clearer overall picture. Is it their enzymes, structure, or something else entirely that controls the hydrogen production? Additionally, in taking microbial hydrogen production to a higher level and larger scale, the strain of bacteria for optimum efficiency must be considered. MC4100 is more expensive and specialized than K-12, so which is more effective will factor into ultimate costs and conditions. Bacterial and algal hydrogen production has generally focused on each in isolation, and algal-bacterial symbiosis is a field lacking true deep understanding.
MATERIALS & METHODS.
Chemicals and vials. Chlamydomonas and E. Coli K-12 were obtained from Carolina, and E. Coli MC4100 was obtained from ATCC. A nanoparticle dispersion of platinum (Pt NP) was also obtained from ATCC. TAP medium was purchased from Fisher-Scientific. Hypo Vials (40mL, septa screw cap) were purchased from Amazon.
Cultivation of pure cultures. For culturing the algae, the culture vial was opened as soon as possible. Then, the cap was loosened, and it was left in normal room lighting until it was time to subculture. The media from Fisher was sterile, but if it was not sterile, it was autoclaved for 15 minutes at 15 pounds of pressure. Then, using a pipette to scrape and wash out algae, 10mL of algae was transferred to 200mL of TAP medium. Then, it was left to grow to an optical density (OD) of 1.4 (750 nm), where it was diluted with another 400 mL of TAP media and left to grow to a final OD of 0.7.
For culturing bacteria, bacteria were transferred to E. Coli to nutrient agar using a cell spreader and left in an incubator for two days at 37°C. Then, it was transferred to 120 mL of TAP medium and cultured and diluted for a final OD of 0.35. The same procedure was used for C. Freundii, except it was left in the incubator at 30°C.
The MC4100 strain had to first be de-thawed and reactivated according to the enclosed instructions. Then, the contents were transferred to a 5-6 mL tube of Luria-Bertani (LB) broth. Additional test tubes and agar plates were inoculated by transferring 0.5 mL of the primary broth to secondary tubes. Several drops of the primary broth were used to inoculate a nutrient agar plate and incubated at 37°C for 24 hours. Finally, the above E.coli procedure was repeated.
Mixed Cultures. For each vial, no headspace was left to ensure as little H2 was able to escape the cultures and the titration was as effective as possible. For the pure algae (PA) category, 40 mL of pure Chlamydomonas culture was put into a vial. For the bacterial cultures, it was split evenly between algae and the particular strain of bacteria,
Analysis via titration. The titration was prepared by combining 16 mL of colloidal platinum (Pt) with 0.3g of methylene blue (MB), dissolved in 99 mL of 95% ethanol; the reagent was dropped into each vial in 1uL drops. The process of titration is depicted in Figures 1 and 2; Figure 1 shows the set-up, tools, and reagent, and Figure 2 shows the color comparison. When the blue color disappeared, the fraction of a 20 uL drop was recorded. To calculate the amount of H2, a conversion factor of 0.29uM/drop was applied [10].
Figure 1. The titration process setup with reagent in a graduated cylinder and micropipettes used for adding reagent dropwise located on the table.20 mL each (E. Coli, Chlamydomonas, and MC4100 mixed cultures named EA, CA, MA, respectively). All experiments were performed with six parallel, identical samples (see Figure S1).
Figure 2. Titration process. A color comparison between a titrated vial (right) and un-titrated vial (left). Done directly under bright light to ensure accuracy in color comparison, though the difference became slim at times.
RESULTS.
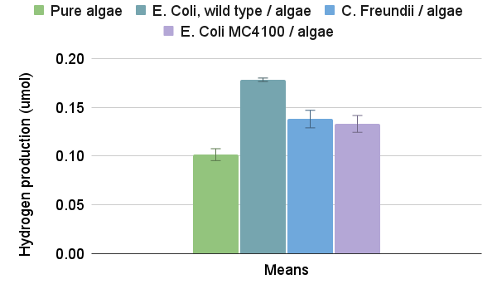
The first three data points in the second (EA) category were omitted from Table 1 (and therefore the dataset for Figure 3) because they were spent figuring out the drop interval, and were thus far too high. Even accounting for that, however, E. Coli K-12 seemed the most effective with a 75.6% increase compared to the control (0.178 umol vs .102 umol), while C. Freundii and MC4100 showed roughly a 30% increase (0.138 umol and 0.133 umol vs 0.102 umol). Otherwise, between the categories, CA and MA showed no statistical difference with overlapping error bars (see Figure 3) and excessively high p values, so it can’t be concluded which is more effective than the other.
| Table 1. Hydrogen production by category by trial (umol) | ||||
| Pure algae | E. Coli, wild type / algae | C. Freundii / algae | E. Coli MC4100 / algae | |
| Trial 1 | 0.102 | 0.160 | 0.131 | |
| Trial 2 | 0.102 | 0.116 | 0.116 | |
| Trial 3 | 0.102 | 0.131 | 0.131 | |
| Trial 4 | 0.087 | 0.174 | 0.087 | 0.145 |
| Trial 5 | 0.102 | 0.189 | 0.145 | 0.131 |
| Trial 6 | 0.116 | 0.174 | 0.189 | 0.145 |
| Means | 0.102 | 0.178 | 0.138 | 0.133 |
Figure 3. Column graph depicting mean hydrogen amounts in umols by category. The error bars indicate standard error per category, taken from the ANOVA. Visually, it’s clear each symbiote had a significant effect, and which were more effective than others.
DISCUSSION.
Following an ANOVA (see Figure S2), these results are significant, and the null hypothesis (that artificial algal-bacterial symbiosis does not affect hydrogen production of algae) can be rejected. Specifically, though, the highest performance of the E. Coli K-12 strain supports the hypothesis and seems to show that having additional hydrogenases is in fact better when choosing an artificial bacterial symbiote. Rather than suffering from detrimental competition for the same resources, the symbiotic culture was greater than the sum of its parts. However, throughout the culturing process, it was clear E. Coli K-12 grew best in the TAP media. It grew to an OD of 0.5, whereas the other two lagged behind and peaked at around 0.3. Proportional results can be observed in both Table 1 and Figure 3, where E. Coli K-12 is clearly leading, while the other two lag behind at about the same rate. A simple but plausible explanation is that E. Coli K-12 is more compatible with Chlamydomonas media and therefore most compatible with Chlamydomonas itself.
All this means there is a simple, cost-effective way of optimizing production of hydrogen using microbial organisms, which has big implications for the larger, potential commercialization of these processes. Though the science of clean energy is a major obstacle, the arguably largest obstacle is investment from both the public and private sector. In using microbes for hydrogen production, there is little for humans to engineer, as nature has already done most of the work in creating these processes; all that remains is how to best exploit them. Governments and corporations alike are hesitant to put time and money into projects that will not see immediate results; however, if those projects are made cheaper and easier, it makes it more enticing to sign legislation, approve investment funds, etc.
Though this project was limited by analysis equipment (titration vs gas chromatography, as would be the standard), it still proved useful and effective. Additionally, it would be beneficial to do research on which aerobic bacteria grow best in TAP media, or the creation of a media that can adequately support both algae and bacteria simultaneously, as that could have been a leading cause in this experiment’s results. Potential future projects include different strains of bacteria/algae, more regulation of the environment to counteract competition or other harmful relationships, and other genetically modified organisms to further enhance efficiency. Since the focus of this project is minimizing costs, it seems best to focus on cheap and well-understood bacteria like C. Freundii and E. Coli K-12 rather than more expensive strains like MC4100 (to put in perspective, the MC4100 strain was over 25 times as expensive), especially since any difference between MC4100’s and C. Freundii’s performances as symbiotes is negligible.
ACKNOWLEDGMENTS.
I’d first like to thank American Heritage School for providing me the space and funding to do this project. Additionally, I’d like to acknowledge Dr. Brittnee McDole and Ms. Jenny Ko, my research teacher and lab advisor. Lots of stressful research and other ordeals went into this project, two years in the making, and I came out on the other side successful thanks to their help. For providing mentorship, time, effort, and expertise to guide me through what I could do and helping me figure out what I couldn’t, thank you.
SUPPORTING INFORMATION.
A Supporting Information document contains an additional image of the experimental process and tables of statistical analysis (ANOVA & Dunnetts test done via excel) and an image.
REFERENCES.
- P. Brandon, Z. Kurban. Clean energy and the hydrogen economy. Philosophical transactions. Series A, Mathematical, physical, and engineering sciences, 375(2098), 20160400 (2017)
- N. Altork, J. R. Busby, Hydrogen fuel cells: part of the solution. Technology & Engineering Teacher, 70(2), 22-27 (2010)
- Ramanan, B.-H. Kim, D.-H. Cho, H.-M. Oh, H.-S. Kim, Algae–bacteria interactions: Evolution, ecology and emerging applications. Biotechnology Advances, 34(1), 14-29 (2016)
- Yao, S. Lyu, Y. An, J. Gjermansen, C. Lu, A. Schram, Microalgae–bacteria symbiosis in microalgal growth and biofuel production: a review. J Appl Microbiol, 126: 359-368 (2019)
- Lakatos, Z. Deak, I. Vass, T. Retflvi, S. Rozgonyi, G. Rakhely, V. Ordog, E. Kondorosi. Bacterial symbionts enhance photo-fermentative hydrogen evolution of Chlamydomonas algae. Green Chem 16, 4716–4727 (2014)
- Wanthanee, R. Rastogib, I. Aran, P. Lindbladd, M. Datta, P. Ashok, C. Larroche, Microbial Production of Hydrogen – A Review. Biosource Technology, 243, 1194-1206 (2017)
- Sharma, S. K. Arya, Hydrogen from algal biomass: A review of production process. Biotechnology reports (Amsterdam, Netherlands), 15, 63–69 (2017)
- UniProtKB – P30131 (HYPF_ECOLI). (n.d.). UniProt. Retrieved June 1, 2022, from https://www.uniprot.org/uniprot/A0A0H3JHT3
- G. Puchenkova, Enterobacteria in areas of water along the Crimean coast. Mikrobiologi Zhurnal (Kyiv, Ukraine). 58(2): 3–7 (1993)
- Seo, R. Kurokawa, B. Sato, A convenient method for determining the concentration of hydrogen in water: use of methylene blue with colloidal platinum. Medical gas research, 2, 1 (2012)
Posted by John Lee on Tuesday, May 30, 2023 in May 2023.
Tags: biochemistry, environmental, hydrogen, microbes, Sustainability