Defining Evaluation Metrics for Medical Imaging Datasets
ABSTRACT
Computer vision (CV) is an application of deep learning that has been gaining increasing significance. CV in healthcare focuses on using medical imaging datasets to train models that help early diagnosis of medical conditions. The model performance relies on the quality of the dataset used. However, there are several limitations of these datasets that hinder their reliability including their availability and size, compromising model performance. This paper defines metrics to evaluate medical imaging datasets to determine a reliable dataset and discusses methods to improve and create such a dataset. The metrics will be implemented and used to evaluate the reliability of three datasets.
INTRODUCTION.
Computer vision (CV), a field under Artificial Intelligence, focuses on enabling computers to identify, classify and segment objects in visual content. CV tasks often rely on neural networks, which can be categorized under supervised and unsupervised learning. Supervised learning refers to networks trained on labelled datasets.
The application of CV in medical imaging, computer-aided diagnosis (CAD), ensures a much faster and accurate diagnosis, early disease recognition, prevents human error, and prevent injuries. CV models in healthcare must have good performance to avoid misclassification and incorrect diagnosis. However, large high quality medical imaging datasets that are needed aren’t easily available. This paper will address the following aspects: (a) Defining metrics to evaluate dataset reliability (b) Methods to improve and create a reliable dataset (c) Evaluation of the reliability of three datasets using the defined metrics
DATA.
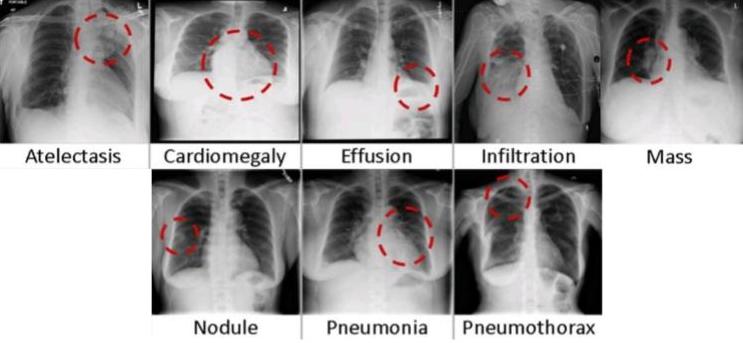
This paper uses three medical imaging datasets. ‘ChestX-ray14’(1) is provided by the National Institute of Health (NIH) Clinical Center. This dataset expands on ‘ChestX-ray8’ (2) with the inclusion of 6 more disease categories. Figure 1 depicts sample images from 8 of the disease classes: Atelectasis, Cardiomegaly, Consolidation, Edema, Effusion, Emphysema, Fibrosis, Hernia, Infiltration, Mass, Nodule, Pleural Thickening, Pneumonia, Pneumothorax.
Figure 1. Sample images from ChestX-ray14 that represent each class (2)
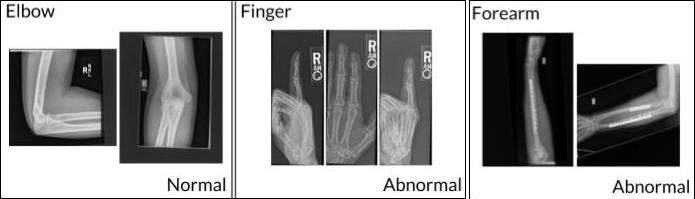
‘MURA (musculoskeletal radiographs)’ (3), is the largest public radiographic image dataset. It was manually labeled by radiologists as either “normal” or “abnormal”. These multi-view radiographic images belong to seven standard study types: elbow, finger, forearm, hand, humerus, shoulder, and wrist, as depicted in Figure 2.
Figure 2. Sample images from MURA that represent each class (3)
CheXpert (4) is a dataset created by a team at Stanford University that includes X-ray images with uncertainty labels. The images represent the following classes: no finding, enlarged cardiom, cariomegaly, lung lesion, lung opacity, edema, consolidation, pneumonia, atelectasis, pneumothorax, pleural effusion, pleural other, fracture and support devices.
METHODS.
- Definition of the metrics. This section defines metrics to evaluate medical imaging datasets. These five metrics consider various aspects of medical imaging datasets to present criteria that will help select a reliable dataset.
1.1. Dataset size. The size of a medical imaging dataset plays a crucial role in the performance of a machine learning model. The size of the data required to train machine learning algorithms varies depending on the use case, the performance level desired, the input features, the algorithm type and architecture, the number of algorithm parameters, and the quality of the training data, including the annotation quality, feature distribution, and noise in the extracted features (5). The rule of 10 is an effective way to determine the dataset size required which states that the sample size needed to train a model should be 10 times the number of parameters. Furthermore, an estimated 150-500 images per class is adequate to prevent both oversampling and under sampling (6).
1.2. Bias. Medical imaging datasets are inherently biased as they don’t reflect the targeted medical condition entirely. As a result, models that perform well on medical imaging datasets often perform poorly when applied to real-world scenarios. Studies (7, 8) have shown that human biases have been both inherited and amplified by artificial intelligence. For instance, facial recognition systems have been proven to perform poorly on underrepresented populations in the datasets based on aspects such as race and gender. A study (9) of the Fitzpatrick Skin Type classification system showed that the datasets (IJB-A and Adience) mainly comprised light-skinned subjects, 79.6% and 86.2% respectively. Furthermore, an analysis and evaluation of gender classification systems revealed that darker-skinned females were the most misclassified group with an error rate of approximately 34.7%. In addition, labeling and annotation error may also lead to dataset bias. Medical imaging datasets are usually labeled by human annotators which may lead to systemic bias in the assigning of labels (10).
1.3. Reliability and annotations. The application of computer vision in healthcare requires high-quality, annotated datasets. Oftentimes, the help of non-experts or automated systems with inadequate supervision are used which increases the unreliability of the dataset (11) Radiologists’, surgical and pathology reports are the ground truth annotation process, other than crowdsourcing annotation, followed. However, these processes aren’t definite depictions of the ground truth (12). For instance, a Mayo Clinic study reported that a major diagnosis was missed clinically in 26% of patients when comparing clinical diagnoses with postmortem autopsies (13).
1.4. Type of data and pixel size. The type of medical imaging data included in the dataset plays a significant role in the computation time. Machine learning models are usually trained on images that have a small matrix size to reduce computation time. However, medical images have a higher dimensionality, ranging from 64 ✕64 to 4000 ✕ 5000. A higher image size indicates a higher number of features that must be extracted which increases computation time.
1.5. Availability of datasets. Private datasets are those that are restricted to only selected individuals or groups of people. Public datasets are those that are freely available. Restricted access to large datasets and high is a major obstacle. In recent years, several large medical imaging datasets such as ChestX-ray14 and MURA have been made available to the public. Computer vision with medical imaging requires large datasets. However, restricted access to these large datasets and high costs are a major obstacle. Datasets made available are often unreliable as they are unstructured with vague usage requirements and incorrect annotations. A study (14) of large medical imaging datasets explored the discrepancies between these datasets and found that the ChestXray14 labels did not adequately reflect the visual content of the pictures, with positive predictive values ranging from 10% to 30% lower than the original documentation values.
- Preprocessing medical images. Data processing is a crucial step that must be undertaken before training a model on a given dataset. In medical imaging preprocessing is mainly followed to enhance image quality and remove noise from the dataset. Preprocessing mainly depends on the use case and the type of images involved.
The use of large medical imaging datasets usually leads to better model performance. These large datasets are not publicly available for all use cases. Preprocessing is usually applied to small datasets of the same use case to increase the dataset size. This is done by applying techniques such as data augmentation that duplicate various versions of each image in a smaller dataset (15)
Medical images are impacted by blurriness, noise, poor contrast, and sharpness that often leads to false diagnosis. Image enhancement techniques (spatial domain enhancement method and frequency domain enhancement method) are applied to improve the quality of medical images and remove noise from the dataset. These methods of image enhancement vary based on the type of medical imaging (16)
- Creating a medical imaging dataset. Large medical imaging datasets are often restricted, have small sample sizes and lack coverage of diverse populations and geographic areas which hinders model performance. Hence, it is important to know how to prepare a reliable medical imaging.
The consent of the respective authorities and subjects must be taken before using any medical data for the development of a dataset. Determining the sampling size and technique by considering its feasibility is a crucial step. The most effective sampling techniques are simple random, systematic, stratified and cluster sampling. In case a large sample size is not feasible, preprocessing techniques such as data augmentation can be implemented to increase the size of the dataset.
After ethical approval and determining the sample size and technique data needs to be accessed and properly de-identified (removal of sensitive/personal data (17). The data must then be transferred to either an external data storage or local data storage.
One of the most crucial aspects of creating a medical imaging dataset is choosing appropriate labels and ground truth definition as most medical image classification models are based on supervised learning approaches. Extracting labels from reports using NLP after obtaining surgical, genomic, pathologic and or clinical outcome data is one of the most scalable approaches to labelling.
EXPERIMENTS & RESULTS.
Table 1 summarizes the results obtained when implementing the metrics defined to the three selected datasets.
| Table 1. Results obtained when using the metrics to evaluate the reliability of the datasets | ||||||
| No. of images | No. of patients | Pixel size | %
Normal cases |
%
Male subjects |
%
Female subjects |
|
| Chest-
Xray14 |
112120 | 30805 | 3000x 2000 | 75 | 56.5 | 43.5 |
| MURA | 40561 | 12173 | 512x 300 | 55 | NA | NA |
| CheXpert | 224316 | 65240 | 390x 320 | NA | 60 | 40 |
The ChestX-ray14 dataset consists of 112,120 frontal chest X-ray scans from 30,805 unique patients, as represented by sample images in Figure 1. This dataset was labeled using NLP and collected from a clinical archive. The number of “normal” or “no finding” images account for approximately 75% of the dataset. A study (14) found that there wasn’t an adequate sample size selected to represent each label in the dataset. The labels of the dataset were evaluated by calculating their positive predictive value (PPV). The labels used to represent image classes were ambiguous, through a visual inspection by a board-certified radiologist. For instance, in the emphysema class most of the cases (86%) had subcutaneous emphysema instead of pulmonary emphysema which resulted in a low PPV value for the class. Another study (18) found that abnormal cases are underrepresented in this dataset (25%). A study (19) of the dataset confirmed that 56.5% of the images (63,340) were of male patients while 43.5% (48,780) were of female patients
The MURA dataset consists of 40,561 images from 14,863 studies and was labeled by board-certified radiologists at the time of clinical interpretation as either ‘normal’ or ‘abnormal’. A study of 100 ‘abnormal’ images found that they consisted of 53 with fractures, 48 with impacted hardware, 35 with degenerative joint disease, and 29 with other abnormalities. The ratio of abnormal to normal cases is 45:55.
The CheXpert dataset consists of 224,316 images from 65,240 patients and was labeled using an automatic rule-based labeler that extracted observations from radiology reports. The tool assigned labels based on the following values: confidence present, absent and uncertainty. The 14 labels of the dataset include “no finding” which refers to images where no pathologies were identified (4). 60% of the images are of male subjects while 40% are of female subjects.
DISCUSSION.
ChestX-ray14 has the highest number of images and subjects included. However, 75% of the images represent normal cases making it biased. As it was labelled with NLP, the error rates of the tool will affect the reliability of the labels (20, 21). To improve the dataset reliability, there should be an equal balance of images in each category. Furthermore, the male to female subject ratio is relatively better than CheXpert resulting in a lower gender bias. MURA has the lowest number of images and subjects included. Techniques such as data augmentation can be applied to increase the dataset size. 55% of the dataset represents normal cases which indicates an equal distribution, avoiding bias in predictions. CheXpert has the highest number of images and subjects included and was labelled using a rule-based automated labeler. One of the main limitations of the CheXpert labeler is that it doesn’t yield probabilistic outputs (22). It has a high gender bias, with a male-female ratio of 60:40. Having an equal balance between male and female subjects will help reduce the gender bias. The datasets are public datasets which often affects the reliability of annotations. For instance, ChestXray14 labels have positive predictive values ranging from 10% to 30% lower than the original documentation values. Figure 3 summarizes the results while also depicting which datasets are the best for each situation based on the metrics.
Figure 3. Summary of results obtained from using the metrics to evaluate the datasets and conclusion.
CONCLUSION.
This paper defined metrics to select a reliable medical imaging dataset and discussed two techniques that can be followed if the criteria are not met. The valuation of the chosen datasets using the metrics defined showed that certain datasets are more ideal for specific use cases. The metrics defined and the suggestions provided to select and create a medical imaging dataset will help improve the accuracy of CAD. One of the main limitations of the metrics defined is that it doesn’t consider anomalies. For instance, models for specific use cases perform better when trained on smaller medical imaging datasets (23). Hence, the defined metrics must consider certain exceptions to the criteria suggested. Furthermore, an extension of this paper can be to investigate the impact of the reliability of a dataset on model performance.
REFERENCES.
- NIH Clinical Center provides one of the largest publicly available chest x-ray datasets to scientific community | National Institutes of Health (NIH), (available at https://www.nih.gov/news-events/news-releases/nih-clinical-center-provides-one-largest-publicly-available-chest-x-ray-datasets-scientific-community) [Accessed: 15-09-22].
- X. Wang, Y. Peng, L. Lu, Z. Lu, M. Bagheri, R. M. Summers, ChestX-ray8: Hospital-scale Chest X-ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases (available at https://uts.nlm.nih.gov/metathesaurus.html) [Accessed: 15-09-22].
- P. Rajpurkar, J. Irvin, A. Bagul, D. Ding, T. Duan, H. Mehta, B. Yang, K. Zhu, D. Laird, R. L. Ball, C. Langlotz, K. Shpanskaya, M. P. Lungren, A. Y. Ng, MURA: Large Dataset for Abnormality Detection in Musculoskeletal Radiographs (available at http://stanfordmlgroup.github.io/competitions/mura.) [Accessed: 15-09-22].
- J. Irvin, P. Rajpurkar, M. Ko, Y. Yu, S. Ciurea-Ilcus, C. Chute, H. Marklund, B. Haghgoo, R. Ball, K. Shpanskaya, J. Seekins, D. A. Mong, S. S. Halabi, J. K. Sandberg, R. Jones, D. B. Larson, C. P. Langlotz, B. N. Patel, M. P. Lungren, A. Y. Ng, CheXpert: A large chest radiograph dataset with uncertainty labels and expert comparison. 33rd AAA31st Innovative Applications of Artificial Intelligence Conference. IAAI 2019, 590–597 (2019).
- M. D. Kohli, R. M. Summers, & J. R. Geis, Medical Image Data and Datasets in the Era of Machine Learning-Whitepaper from the 2016 C-MIMI Meeting Dataset Session. J. Digit. Imag. 30, 392-399 (2017).
- S. Shahinfar, P. Meek, G. Falzon, “How many images do I need?” Understanding how sample size per class affects deep learning model performance metrics for balanced de-signs in autonomous wildlife monitoring. Ecological Informatics. 57, 101085 (2020).
- T. Bolukbasi, K.-W. Chang, J. Y. Zou, V. Saligrama, A. T. Kalai, Man is to Computer Programmer as Woman is to Homemaker? Debiasing Word Embeddings. 30th Conference on Neural Information Processing Systems. NIPS 2016, 4356–4364 (2016).
- J. Zou, L. Schiebinger, AI can be sexist and racist — it’s time to make it fair. Nature. 559, 324-326 (2018).
- J. Buolamwini, T. Gebru, Gender Shades: Intersectional Accuracy Disparities in Commercial Gender Classification. Conference on Fairness, Accountability, and Transparency. 81, 77-91 (2018).
- L. Joskowicz, D. Cohen, N. Caplan, J. Sosna, Inter-observer variability of manual contour delineation of structures in CT. European Radiology. 29, 1391–1399 (2019).
- D. Karimi, H. Dou, S. K. Warfield, A. Gholipour, Deep learning with noisy labels: exploring techniques and remedies in medical image analysis. Medical Image Analysis. 65, 101759 (2020).
- P. Goddard, A. Leslie, A. Jones, C. Wakeley, J. Kabala, Error in radiology. The British Journal of Radiology. 74, 949-951 (2014).
- J. Roosen, E. Frans, A. Wilmer, D. C. Knockaert, H. Bobbaers, Comparison of premortem clinical diagnoses in critically iII patients and subsequent autopsy findings. Mayo Clinic Proc. 75, 562–567 (2000).
- L. Oakden-Rayner, Exploring Large-scale Public Medical Image Datasets. Academic Radiology. 27, 106–112 (2020).
- P. Bhuse, B. Singh, P. Raut, Effect of Data Augmentation on the Accuracy of Convolutional Neural Networks. Information and Communication Technology for Competitive Strategies. 191, 337–348 (2021).
- S. Patil, V. R. Udupi, Preprocessing to Be Considered for MR and CT Images Containing Tumors. IOSR Journal of Electrical and Electronics Engineering (IOSRJEEE). 1, 54–57.
- M. J. Willemink, W. A. Koszek, C. Hardell, J. Wu, D. Fleischmann, H. Harvey, L. R. Folio, R. M. Summers, D. L. Rubin, M. P. Lungren, Preparing Medical Imaging Data for Machine Learning. Radiology. 295, 4-15 (2020).
- P. Harzig, Y.-Y. Chen, F. Chen, R. Lienhart, Addressing Data Bias Problems for Chest X-ray Image Report Generation. British Machine Vision Conference Press. BMVC 2019, 1-11 (2019).
- G. Stanovsky, N. A. Smith, L. Zettlemoyer, Evaluating gender bias in machine translation. Association for Computational Linguistics. 117, 1679–1684 (2020).
- A. Casey, E. Davidson, M. Poon, H. Dong, D. Duma, A. Grivas, C. Grover, V. Suárez-Paniagua, R. Tobin, W. Whiteley, H. Wu, B. Alex, A systematic review of natural language processing applied to radiology reports. BMC Medical Inform. and Decis. Mak. 21, 1–18 (2021).
- T. Ly, C. Pamer, O. Dang, S. Brajovic, S. Haider, T. Botsis, D. Milward, A. Winter, S. Lu, R. Ball, Evaluation of Natural Language Processing (NLP) systems to annotate drug product labeling with MedDRA terminology. J. Biomed. Inform. 83, 73–86 (2018).
- M. B. A. M. McDermott, T. M. H. H. Hsu, W.-H. Weng, M. Ghassemi, P. Szolovits, CheXpert++: Approximating the CheXpert Labeler for Speed, Differentiability, and Probabilistic Output CSAIL. Proceedings of Machine Learning Research. PMLR 2020, 1-14 (2020).
- A. Khan, M. Usman, S. Zulfikar, A. Bhutto, Alzheimer’s Disease Prediction Model Using Demographics and Categorical Data. iJOE. 15, 96–109 (2019).
Posted by John Lee on Tuesday, May 30, 2023 in May 2023.
Tags: Computer Vision, Dataset Reliability, Medical Imaging Datasets