Comparative Analysis of Prostate Cancer Detection Methods in Emulating Ground Truth Accuracy
ABSTRACT
Segmentation for prostate cancer, the most diagnosed cancer among American men, has become increasingly important for treatment planning. Ground truth, radiologist-annotated data is scarce and thus calls for a viable alternative to costly manual segmentation using deep learning networks. Given the rise in variability and diversity in machine learning models, it is necessary to holistically assess the performance of the three mainstream method groups: supervised, unsupervised, and semi-supervised learning. The survey provides a synopsis of deep learning frameworks and their best-fit image processing applications by comparatively analyzing the efficacy of nine deep learning algorithms for prostate cancer segmentation. While all three algorithm groups could effectively be employed to complete segmentation, it was found that supervised learning is optimal for small-scale classification problems with a plethora of annotated data, and unsupervised learning is optimal for feature extraction prior to diagnosis. Semi-supervised learning, a middle ground between the two, has been found to be versatile and conducive to a lack of annotated data, making it the most viable solution for prostate cancer segmentation.
INTRODUCTION.
Prostate cancer (PCa) segmentation is the identification of spatial locations of prostate tumors in a slice-by-slice manner. As PCa has risen to become the most commonly diagnosed cancer among American men [1], it has become vital to identify tumor regions accurately in early stages to prevent further aggravation and facilitate early treatment planning. While manual segmentation is appealing to achieve reliable diagnoses for disease prediction, its viability is limited by the high cost of ground truth labels.
The increasing cost of carrying out manual diagnosis has effected an increase in deep learning algorithms that aid the diagnosis process of clinicians. To transition these various deep learning algorithms into practice, their efficacy and reliability must first be evaluated. Deep learning is an alternative to manual data analysis and image processing that relies on a multi-layered architecture to train on input data and provide predictions without a large compromise in accuracy. DL involves learning from multiple layers of data to achieve meaningful insights in data and make intelligent decisions, which renders it one of the most successful tools for automating tasks requiring human critical thinking, such as prostate cancer diagnosis [2]. The variety of emerging deep learning frameworks brings about the query: Which deep learning approach for prostate cancer segmentation (supervised, unsupervised, and semi-supervised) can best mimic the accuracy of ground-truth annotations? The survey assesses diverse prostate cancer detection algorithms to gauge their effectiveness for clinical applications.
Deep Learning. Machine learning is a subset of artificial intelligence where computers learn from data to perform intuitive tasks. Deep learning, a subset of machine learning, is composed of a series of hidden layers that detect connections in data by emulating the behavior of human neurons [3]. Deep learning can be implemented in settings that involve complex predictions or the classification of distinct groups. Deep learning algorithms have been implemented to address a wide range of medical problem areas. For instance, one study trains a model using deep learning to analyze and identify hidden trends in skin cancer images and achieved an AUC of 99.77% in detecting cancer cells from input images [4]. Thus, deep learning has proved itself as an extremely potent tool within the medical diagnosis realm.
Ground Truth Annotations. A ground truth serves as a comparison source for new mechanisms that aim to complete the same task [5]. Ground truth data for PCa comprises diagnoses prepared by human clinicians and is costly due to the manual labor involved. Ground truth samples may include prostate image scans annotated to indicate regions of abnormalities or a clinical diagnosis given a set of patient risk factors. Medical professionals must spend extensive time manually verifying diagnoses to provide accurate suggestions for further clinical treatment for patients. Also, specialized medical professionals are limited in availability, which contributes to the scarcity of ground truth data. Segmentation networks should reproduce clinical diagnoses on scarce training data to mitigate heavy ground truth costs [6].
Automated Segmentation. Segmentation consists of dividing an input into unlabeled or pre-categorized labels [7]. Threshold-based segmentation assigns a threshold value to each individual pixel used to create a binary mask of the original image [8]. Edge-based segmentation examines regions of discontinuities in gray levels and vibrancies in an image to determine preliminary borders; then, these disconnected borders undergo another layer of segmentation to create a more seamless segmentation result [9]. Medical image segmentation can be applied on several image modalities, the two most prominent being magnetic resonance (MR) imaging and computed tomography (CT) imaging [10]. MR imaging is conducive to segmentation as it features a high resolution and high signal to noise ratio. Models that achieve high-accuracy segmentation with minimal false positive and false negative classifications may be considered for clinical implementation as an alternative to cost-heavy manual diagnosis approaches.
Learning Techniques. Supervised learning is a subset of machine learning which learns from labeled pairs in ground truth data [11]. One technique, binary classification, can produce a value between 0 and 1 for each pixel to indicate the probability of the pixel being cancerous. A common supervised framework splits the data into training, test, and validation sets. The algorithm learns from input-output pairs and creates a hierarchy of learned information through feedback from a loss function [12]. Unsupervised learning embodies a free-form approach to pattern recognition. It is more receptive to subtler, hidden trends in data and has the capability to draw predictions or identify clusters without the limitations of output bounding boxes [13]. Unsupervised models place emphasis on finding deep-level trends in data and thus employ dimension reduction techniques such as principal component analysis to keep sight of the most relevant key features [14]. Semi-supervised learning draws upon both supervised and unsupervised techniques, by training a model upon subsets of labeled and unlabeled data, and it has increased in popularity for its versatility [15]. Semi-supervised models rely one or more assumptions. The continuity assumption states that points close in proximity are likely to share a label. The manifold assumption states that the data points lie on a low dimensional manifold within a higher dimensional space. The cluster assumption states that data usually form distinct clusters that share a common label [16].
Overview of Efficiency Metrics. F-measure metrics measure the accuracy of the prediction with regard to its overlap with the ground truth. The Jaccard index, also known as the Intersection-over-Union (IoU) measures the area of overlap between the target bounding box and the prediction. The Dice similarity coefficient (DSC) is the harmonic mean between sensitivity and specificity. Sensitivity is the number of true positives over the total number of true positives and false negatives. Specificity is the total number of true negatives over the total number of true negatives and false positives. Accuracy is a commonly used metric for general use but may produce misleading results for medical image segmentation: if the area of interest is small but the algorithm labels all pixels as background pixels, accuracy would be high regardless of the number of incorrect pixels. The Receiver Operating Characteristic (ROC) curve graphs the true positive rate against the false positive rate. It is a performance indicator of a single-value binary classifier that provides a value from 0.5 to 1, where 0.5 is the performance of a random classifier, and 1 is the performance of a perfect classifier [17][18].
MATERIALS AND METHODS.
| Table 1. Summary of nine methods used for comparative analysis, including method name, method structure, efficiency metric used for testing, and modality of dataset images. | |||
| Method | Structure | Metric | Modality |
| Logistic Regression [19] | Supervised | F1 Scoring; Accuracy | CV |
| SPCNet [20] | Supervised | AUC ROC | MRI |
| Deep Learning Segmentation [21] | Supervised | AUC ROC | Multi-parametric MRI |
| UATS [22] | Semi- Supervised | DSC | Multi-parametric MRI |
| Deep Learning with End-to-end Streaming [23] | Semi- Supervised | AUC ROC | Prostate Biopsy |
| Dual Architecture Model [24] | Semi- Supervised | AUC ROC | Biparametric MRI |
| SCO-SSL [25] | Semi- Supervised | DSC | Transrectal Ultrasound (TRUC) |
| DeepT2Vec [26] | Unsupervised | F1 Scoring; Accuracy | Transcriptomic tissue and tumor data |
| AI-Biopsy [27] | Unsupervised | AUC ROC; Accuracy | MRI |
Supervised Methods.
(i) Logistic Regression [19]: A study conducted by Mahamudul Hasan et al., assesses various supervised machine learning techniques for binary prostate cancer classification. The study compares six supervised machine learning techniques to predict a patient’s diagnosis for prostate cancer (0 or 1) based on 100 patient records on 10 distinct quantitative risk factors, including radius, texture, perimeter, area, and smoothness. The dataset, from a Kaggle repository, includes 62 cancer-positive records and 38 non-cancerous records. The model used a train-test split of 80-20 and was built on a simple framework of preprocessing, data analysis, splitting, inputting into a classifier model, and finally prediction.
(ii) Convolutional Neural Network Architecture [20]: A study conducted by Arun Seetharaman et al., features a custom-built neural network architecture called the Stanford Prostate Cancer Network (SPCNet), which is trained to differentiate between aggressive, indolent, and normal tissues on a pixel-by-pixel basis on a prostate cancer MRI image. The model architecture features four convolution layers, with a final SoftMax layer, followed by two ReLU activation layers. The final result is upsampled and annotated on a mask with key colors representing different output classes.
(iii) Retina U-Net Framework Deep Learning Model [21]: Oscar Pellicer-Valero et al. proposes a deep learning pipeline based on the Retina U-Net model for the segmentation and Gleason grade stratification of prostate cancer lesions from multiparametric MRI images. The model was trained on a series of five cascading CNNs. These CNNs make up a broader U-Net architecture that produce a prostate segmentation mask from a T2 scan, produce a Central Gland (CG) mask from the T2 scan and the previous output, and compute the Peripheral Zone (PZ) mask, respectively. A Bounding Box regressor and classifier are layered on top and extract a feature map from the decoder portion to perform classification.
Semi-Supervised Methods
(i) Uncertainty-Guided Self Learning Model [22]: A deep learning semi-supervised architecture created by Anneke Meyer et al., combines temporal ensembling and self-learning to create an uncertainty-aware temporal self-learning model (UATS). The architecture is built around two premises: self-training and self-ensembling. A self-training model generally relies on the cluster assumption, and that confident predictions are correct. As the model progresses, training data is accumulated. However, self-learning does not include a verifying mechanism for early mistakes which may be detrimental to the long-term performance of the model.
(ii) End-to-End Convolution Streaming Architecture [23]: Hans Pinckaers et al., propose an end-to-end deep learning CNN implementation based in ResNet-34 that extracts meaningful features while delivering high-resolution images to work around the scarcity of ground truth data. The model utilized ImageNet-trained weights and SGD as a learning optimizer. A memory-efficient technique of streaming reconstructed a feature map mid-network to optimize GPU memory space and proceed through the neural network.
(iii) DA-UNet and nnU-Net Deep Learning Framework [24]: A study conducted by Joeran Bosma et al., proposes a semi-supervised approach that pairs unlabeled data with clinical reports to optimize data. The model utilizes two distinct architectures based in nnU-Net and Dual-Attention U-Net which are self-selected according to the input dataset. The nnU-Net architecture was trained using a combination as cross-entropy and soft Dice as losses, while the latter was trained using Focal Loss. During segmentation, the number of csPCa lesions and the PI-RADS score were extracted.
(iv) Novel Shadow-Consistent Deep Learning Mechanism [25]: Research conducted by Xuanang Xu et al., focuses on the importance of efficient prostate segmentation in the transrectal ultrasound (TRUS) modality. The algorithm implements shadow augmentation and shadow dropout that involves training samples with augmented shadow patterns to make the algorithm more robust and less sensitive to such added effects. The model was trained in both semi-supervised and supervised settings on large TRUS clinical data.
Unsupervised Methods
(i) Deep Learning Autoencoder [26]: The lack of similar features or correlation between transcriptions led Bo Yuan et al. to construct a deep unsupervised network to extract high-level features from human tissue and tumor samples. Five layers with successively decreasing neurons were trained in an unsupervised manner for feature extraction and conversion into low dimensional vector data known as a transcriptomic feature vector (TFV). The researchers trained the model on Pan-Cancer samples with both grading and staging information and analyzed each sample and its TFV on a class-level. A semi-supervised architecture was created to differentiate Pan-Cancer and regular classification with a SoftMax classifier.
(ii) Deep Neural Network Using Class Activation Maps (CAM) [27]: The authors of the study, Pegah Khosravi et al., use a Prostate Imaging Reporting and Data System (PI-RADS) for the interpretation of prostate images and to facilitate detection, treatment suggestion, and risk stratification. A novel unsupervised deep learning algorithm using ResNet-152 was trained on ImageNet to first reduce the dimensionality of feature vectors and then extract uninformative and informative feature vectors to isolate the main cluster. A Gaussian Mixture Model (GMM) was then applied for optimal conservation.
DISCUSSION.
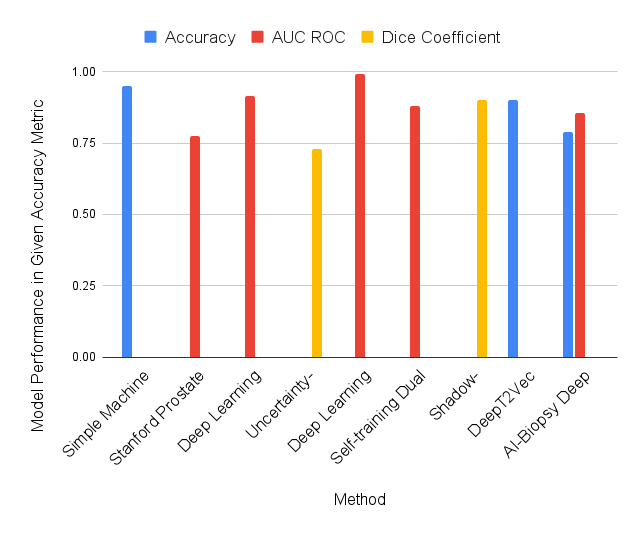
Figure 1 indicates model performance across the three method groups. The logistic regression model [19] achieved an accuracy of 95%, precision of 96%, recall of 95%, and F1 score of 95%. However, the training dataset featured only 100 population samples, which is not representative. The model was trained on CV data rather than 2D MRI images, which does provide output as comprehensive as segmentation programs. The SPCNet Convolutional Neural Network Model [20] was able to catch missed lesions by radiologists and segment aggressive and indolent cancers, which is a notable given the lack of ground truth data. SPCNet can further fine-tune its capability for insight if trained on data that includes additional biometric factors, such as gland size and biopsy status, to more closely tailor its predictions to real-world radiologist decision-making. The Deep Learning Segmentation model [21] achieved an overall AUC ROC of 0.96. The algorithm’s compatibility with mpMRI prostate scans puts it at a slight disadvantage because these lesions are small with ill-defined margins, thus leading to a high variability for interpretation. The structure of all three algorithms have room for improvement; SPCNet can benefit from the incorporation of clinical factors into decision making, and the deep learning segmentation algorithm’s structure could be optimized to be competitive with the mentioned simpler ROI classification systems. Supervised learning is a tool best suited for classification problems.
Figure 1. Performance of nine methods across three method categories (supervised, semi-supervised, unsupervised), measured in the indicated efficiency metric
The uncertainty-aware self-learning architecture [22] achieved an average DSC score of 73% across all prostate zones. While this score cannot be directly compared with the scores of other algorithms tested with different metrics in this study, the algorithm can be compared to other algorithms in the ISIC2018 skin lesion segmentation challenge using the same dataset. Under these testing conditions, it achieved a DSC of 86%, which was 6.2% below the leading algorithm. However, this may have been attributed to discrepancies in training data size and intent of the algorithm structure. Semi-supervision significantly improves output quality when there is a lack of viable training data, but the gap in quality diminishes as availability of training material increases. The end-to-end CNN model [23] achieved an AUC ROC of 0.992 through streaming and 0.990 through multiple instance learning. The streaming process made it possible for convolutional neural networks to be rapidly trained with high-resolution labeled images, making it competitive with patch-based deep learning techniques. The self-training dual architecture model [24] performed well for both UNet variations, with a 0.894 AUC ROC for nnU-Net and a 0.873 AUC ROC for DA-UNet. Semi-supervised learning displayed a significant improvement in AUROC performance compared to supervised training on a test set of 2,440 manual exams. The semi-supervised nature of the model allowed for rapid automatic labeling for new cases. The model was drastically improved by applying semi-supervised learning to training but exhibits model-specific limitations such as narrow training data. The shadow-consistent deep learning model [25], achieved a DSC of 92.44% for one population dataset and a score of 87.98% for a second population, with both models implementing UNet. It showed significant improvement as the number of labeled samples decreased, and thus proves to be applicable in the prostate cancer segmentation realm where ground truth data is highly limited.
The unsupervised deep autoencoder for feature extraction [26] could pull key features into TFVs of transcriptomic data. The portion of the algorithm involving unsupervised learning was responsible for the identification of biological features in the data; when connected to the supervised, cancer diagnosis portion of the model, it yielded better results. The AI Biopsy model [27] was evaluated on both AUC ROC and accuracy; it was able to achieve an AUROC of 0.855 and an accuracy of 79.02%. The algorithm utilizes holistic analysis of the MRI image and thus leverages all available data; also, the algorithm does not rely on preprocessing augmentations for new images [28]. However, the histopathologic data used for this algorithm still has its limitations regarding mislabeling.
CONCLUSION.
Supervised learning is a viable option to achieve high accuracy for classification problems with a plethora of labeled data; however, ground truth data for prostate cancer segmentation is scarce. Supervised algorithm performance due to a low signal to noise ratio can be ameliorated by employing a technique such as Principal Component Analysis to discriminate a lesion more effectively from background noise. Moreover, to address the scarcity of available data, data augmentation and auto encoders can be utilized to increase training data by generating similar data points. Semi-supervised learning is arguably the most effective technique for prostate cancer segmentation, as it is conducive to a lack of annotated training data and versatile algorithm structures, which are optimal for this area of image processing. Unsupervised learning may not be the most effective solution for PCa segmentation, which benefits from a classification mechanism training on a structured, supervised regimen. Future study may include reviewing a broader range of existing deep learning techniques and architectures to seek out which general approaches are best suited to prostate cancer diagnosis and paint a more complete picture of the efficacy of current algorithms. In addition, a study can hone in on individual algorithms and remedy their shortcomings by using techniques – namely Principal Component Analysis and Independent Component Analysis – to segregate ill-defined lesions more accurately. Certain models, when training on limited data, may also be prone to overfitting (overly specific model) or underfitting (inadequately trained), which can be explored more in-depth. Also, while deep learning for PCa has garnered popularity, machine learning algorithms, including gradient boosted trees, should be evaluated for their performance against DL algorithms. The study can be taken to a more universal context by expanding its domain to other carcinomas and image modalities.
ACKNOWLEDGMENTS.
Thank you for the guidance of Hina Ajmal from Oxford University in the development of this research paper.
REFERENCES.
- Aldoj, N., Biavati, F., Michallek, F. et al. Automatic prostate and prostate zones segmentation of magnetic resonance images using DenseNet-like U-net. Sci Rep 10, 14315 (2020).
- Saria, S., Butte, A., Sheikh, A. Better medicine through machine learning: What’s real, and what’s artificial? PLoS Med. 15(12), e1002721 (2018).
- Sarker, I. H. Deep Learning: A Comprehensive Overview on Techniques, Taxonomy, Applications and Research Directions. SN Comput Sci. 2(6), 420 (2021).
- Kadampur, M., Al Riyaee, S. Skin cancer detection: Applying a deep learning based model driven architecture in the cloud for classifying dermal cell images. Informatics in Medicine Unlocked 18 100282 (2020).
- Cardoso, J. R., Pereira, L. M., Iversen, M. D., et al. What is gold standard and what is ground truth? Dental Press J Orthod. 19(5), 27-30 (2014).
- Li, J., Udupa, J., Tong, Y., et al. Segmentation evaluation with sparse ground truth data: Simulating true segmentations as perfect/imperfect as those generated by humans. Medical Image Analysis 69, 101980 (2021).
- Hesamian, M. H., Jia, W., He, X., et al. Deep Learning Techniques for Medical Image Segmentation: Achievements and Challenges. J Digit Imaging. 32, 582-596 (2019).
- Wu, D., Yuan, C. Threshold image segmentation based on improved sparrow search algorithm. Multimed Tools Appl. (2022).
- Fan, S. L. X., Man, Z., Samur, R. Edge based region growing: a new image segmentation method. Association for Computing Machinery, 302-305 (2004).
- Sharma, N., Aggarwal, L. M. Automated medical image segmentation techniques. J Med Phys. 35(1), 3-14 (2010).
- Jiang, T., Gradus, J. L., Rosellini, A. J. Supervised Machine Learning: A Brief Primer. Behav Ther. 51(5), 675-687 (2020).
- Le’Clerc Arrastia, J., Heilenkötter, N., Otero Baguer, D., et al. Deeply Supervised UNet for Semantic Segmentation to Assist Dermatopathological Assessment of Basal Cell Carcinoma. J Imaging. 7(4), 71 (2021).
- Aganj, I., Harisinghani, M.G., Weissleder, R. et al. Unsupervised Medical Image Segmentation Based on the Local Center of Mass. Scientific Reports 8, 13012 (2018).
- Kim, W., Kanezaki, A., Tanaka, M. Unsupervised Learning of Image Segmentation Based on Differentiable Feature Clustering. IEEE Transactions on Image Processing29, 8055-8068 (2020).
- Van Engelen, J.E., Hoos, H.H. A survey on semi-supervised learning. Machine Learning 109, 373-440 (2020).
- Li, Y., Liang, D. Safe semi-supervised learning: a brief introduction. Frontiers of Computer Science 13, 669-676 (2019).
- Nai, Y., Teo, B.W., Tan, N.L., et al. Comparison of metrics for the evaluation of medical segmentations using prostate MRI dataset. Computers in Biology and Medicine134, 104497 (2021).
- Melo, F. Area under the ROC Curve. In: Dubitzky, W., Wolkenhauer, O., Cho, K.H., Yokota, H. Encyclopedia of Systems Biology. Springer, New York, NY (2013).
- Hasan, S.M., Rabbi, M.F., Jahan, N. Can Machine Learning Technique Predict the Prostate Cancer accurately?: The fact and remedy. In: 2021 International Conference on Electronics, Communications and Information Technology (ICECIT), pp. 1-4 (2021).
- Seetharaman, A., Bhattacharya, I., Chen, L.C., et al. Automated detection of aggressive and indolent prostate cancer on magnetic resonance imaging. Medical Physics 48, 2960-2972 (2021).
- Pellicer-Valero, O.J., Marenco Jiménez, J.L., Gonzalez-Perez, V., et al. Deep learning for fully automatic detection, segmentation, and Gleason grade estimation of prostate cancer in multiparametric magnetic resonance images. Scientific Reports 12, 2975 (2022).
- Meyer, A., Ghosh, S., Schindele, D., et al. Uncertainty-aware temporal self-learning (UATS): Semi-supervised Learning for segmentation of prostate zones and beyond. Artificial Intelligence in Medicine 116, 102073 (2021).
- Pinckaers, H., Bulten, W., van der Laak, J., et al. Detection of prostate cancer in whole-slide images through end-to-end training with image-level labels. IEEE Transactions on Medical Imaging, 40(7), 1817-1826 (2021).
- Bosma, J. S., Saha, A., Hosseinzadeh, M., et al. Annotation-efficient cancer detection with report-guided lesion annotation for deep learning-based prostate cancer detection in bpMRI. https://doi.org/10.48550/arXiv.2112.05151 (2021).
- Xu, X., Sanford, T., Turkbey, B., et al. Shadow-consistent semi-supervised learning for prostate ultrasound segmentation. IEEE Transactions on Medical Imaging, 41(6), 1331-1345 (2022).
- Yuan, B., Yang, D., Rothberg, B. E. G., et al. Unsupervised and supervised learning with neural network for human transcriptome analysis and cancer diagnosis. Scientific Reports, 10, 19106 (2020).
- Khosravi, P., Lysandrou, M., Eljalby, M., et al. Biopsy-free prediction of prostate cancer aggressiveness using deep learning and radiology imaging. Journal of Magnetic Resonance Imaging. (2019)
- Brown, J. B. Classifiers and their metrics quantified. Molecular Informatics, 37(1-2), 1700127 (2018).
Posted by John Lee on Tuesday, May 30, 2023 in May 2023.
Tags: Deep learning, MRI images, prostate cancer