A Review and Comparative Analysis of Breast Cancer Segmentation Using U-Net and Its Variants
ABSTRACT
Breast cancer is the most diagnosed type of cancer in women. The manual process of histopathological analysis is laborious, time-consuming, and limited by the quality of the specimen and the experience of the pathologist. Accurate segmentation of the cancerous cells is still a challenging task. In this study, the objective is to conduct an evaluation of the U-net and its variants (Attention U-net and Residual Attention U-net) on a set of histopathological images 2018 Data Science Bowl and provide a comparative analysis of the results. All the methods are evaluated using accuracy and the results of the networks trained on the dataset demonstrated that using Residual Attention U-net, breast cancer can be accurately segmented.
INTRODUCTION.
Breast Cancer is the most diagnosed type of cancer. It is the second most common cause of cancer-related death in women.[1] Accurate diagnosis, staging and grading require tissue biopsies. These biopsies must be processed onto slides and stained. Hematoxylin and Eosin are two common stains used to identify cytoplasm and nuclei. The hematoxylin stains the nucleus blue, while the Eosin stains the cytoplasm pink. Examination of these tissue biopsies is currently considered the gold standard for clinical diagnosis of cancer.[2]
In recent times, the field of digital pathology has been made possible with the introduction of whole slide image (WSI) scanners [3] and has further been driven by image technologies, data storage, and computer vision algorithms [4]. The WSIs allow for in silico analysis. An in-silico experiment is one performed on a computer or via computer simulation. The application of artificial intelligence (AI) in silico analysis is proving to be a promising tool for digital pathology. [4]
Researchers and data scientists in AI have applied convolutional neural networks (CNN) to segment and classify objects, predict diseases, and therefore treatment responses [4]. Recent developments in CNN architectures show the rapid development in the field of deep learning [5]. The CNNs trained on large, supervised image datasets can carry out classification and segmentation tasks for medical image analysis. [6]
U-Net is an example of a deep convolutional neural network (DCNN) that provides semantic segmentation. The network was first developed by Ronneberger el at., building on the fully convolutional network (FCN) [7]. U-Net improved on the FCN in multiple ways such as reducing training time, enhancing localization with the addition of long skip connections, and up-sampling layers to the encoder-decoder architecture. [8]-[11]
Although Unet has good representational power, it uses multiple skip connection that bring redundant low-level features from the lower level layers. Novel attention gate (AG) model implicitly learns to suppress irrelevant regions in an input image while highlighting salient features useful for a specific task. AG models for medical imaging automatically learns to focus on target structures of varying shapes and sizes. An implementation of AG in a standard U-Net architecture (Attention U-Net) and apply it to medical images. [12].
Residual U-net provided a solution to the problem by adding skip connections that help preserve the feature maps while going deeper in the network and retains the performance for such networks. Residual attention U-net adds the attention module to the residual U-net that helps to capture the multilevel features in the global context. Overall, the residual attention U-net works better with higher resolution images that provide more details of the location information and performs better without going deeper in the network. [13]-[14]
Therefore, this study aims to compare the three deep convolutional neural networks: U-Net, Attention U-Net, and Attention Res-Net. In the methodology sections, we describe the dataset and an overview of the architecture of the selected methods. In the results and experimentation section, we give the details of our experimental setup and provide a comparative analysis of the results.
MATERIALS AND METHODS.
Dataset. The dataset used to compare the three deep convolutional neural networks is the 2018 Data Science Bowl. The images used were acquired under various conditions and varied in cell type, magnification, and imaging. The dataset contains 670 segmented nuclei. Each image in the dataset is associated with its own unique ImageID. The files belonging to an image are contained in a folder with this ImageID. Within the folder, there are two subfolders, one named images which contains the image file, and another file names masks which contains the segmented masks of each nucleus. The original dataset has one or multiple masks depending on how many nuclei are there in an image. Each mask has one nucleus. For convenience, we merged all masks and generated a single mask against each image. The size of each image is 256X256 pixels.
The U-Net algorithm will require the following file structure for the training data: The main folder which is named its own unique ImageID. Under the main folder, there are two subfolders: Images and masks. The image folder has an image with all the nuclei, with the image name being its own unique ImageID. The masks folder contains one mask having all nuclei for each image.
The Attention U-Net algorithm and Attention Res-Net will require the following file structure: images and masks. Under the images folder, images with its own ImageID. The masks folder contains the masks for the images, which were formed by combining all the masks from their masks folder.
Overview of the methods. All Three of the algorithms are fully Convolutional Networks (FCN). Convolutional neural networks (CNNs) outperform traditional approaches in medical image analysis on public benchmark datasets,[15]-[16] while being an order of magnitude faster than.[17] This is mainly attributed to the fact that (1) domain-specific image features are learnt using stochastic gradient descent (SGD) optimization, (2) learnt kernels are shared across all pixels, and (3) image convolution operations exploit the structural information in medical images well. Fully convolutional networks (FCN) [18] such as U-Net [19] have been shown to achieve robust and accurate performance in various tasks including cardiac MR [20] and brain tumors [21] image segmentation tasks.
U-Net. U-Net is the first encoder-decoder set up for medical image segmentation. There are two important parts of the U-net, the encoder and the decoder. Each block in the contracting path consists of two successive 3×3 convolutions followed by a ReLU activation unit and a max-pooling layer. This arrangement is repeated several times. The novelty of U-net comes in the expansive path where at each stage the feature map is up-sampled using 2×2 up-convolution. Then, the feature map from the corresponding layer in the contracting path is cropped and concatenated onto the up-sampled feature map. This is followed by two successive 3×3 convolutions and ReLU activation. At the final stage, an additional 1×1 convolution is applied to reduce the feature map to the required number of channels and produce the segmented image. The cropping is necessary since pixel features in the edges have the least amount of contextual information and therefore need to be discarded.
Attention U-Net. An often-desirable trait in an image processing network is the ability to focus on specific objects that are of importance while ignoring unnecessary area[12]. The attention U-net achieves this by making use of the attention gate [22]. An attention gate is a unit which trims features that are not relevant to the ongoing task. Each layer in the expansive path has an attention gate through which the corresponding features from the contracting path must pass through before the features are concatenated with the up-sampled features in the expansive path. Repeated uses of the attention gate after each layer improves segmentation performance significantly without adding too much computational complexity to the model.
The attention unit is useful in encoder-decoder models such as the U-net since it can provide localized classification information as opposed to global classification. In U-net this allows different parts of the network to focus on segmenting different objects. Furthermore, with properly labeled training data, the network can attune to particular objects in an image. The attention gate applies a function where the feature map is weighted according to each class and the network can be tuned to focus on a particular class [23], and hence pay attention to particular objects in an image. While there are different types of attention gates, additive attention is more popular in image processing due to it resulting in higher accuracy.
Attention Res-Net. This variant of U-net is based on the ResNet [24] architecture. The motivation behind ResNet was to overcome the difficulty in training highly deep neural networks. There is evidence in the literature that increasing the number of layers results in saturation, and further increases can cause degradation of performance [25]. This degradation arises due to the loss of feature identities in deeper neural networks caused by diminishing gradients in the weight vector. ResNet lessens this problem by utilizing skip connections which take the feature map from one layer and add it to another layer deeper in the network. This behavior allows the network to better preserve feature maps in deeper neural networks and provide improved performance for such deeper networks.
Residual Attention U-Net (RAUNet) adopts an encoder-decoder architecture to get high-resolution masks. The architecture of RAUNet is pre-trained on ImageNet and is used as the encoder to extract semantic features. It helps reduce the model size and improve inference speed. In the decoder, a new attention module augmented attention module (AAM) is designed to fuse multi-level features and capture global context. Furthermore, transposed convolution is used to carry out up-sampling for acquiring refined edges. The Residual Attention U-net provides a solution which can prevent the network from going very deeper but retains the performance.
RESULTS AND DISCUSSION.
All software for this study was written in the Python programming language. The Attention U-Net uses TensorFlow version 1.5.0, Keras version 2.0.8. Both U-Net and Attention Res-Net use TensorFlow version 2.9.1, Keras version 2.9.0, Scikit-image version 2.9.0, cv2 version 4.6.0.66 and scikit-learn version 1.1.2
There are 9 layers in the model with a filter size of 3×3. Each of the layers in the deep learning neural network used activation of ‘ReLU’, a kernal_initialzer of ‘he_normal’, and padding of ‘same’. The model takes the input height, width and number of channels as the input. All the images fed into the network have a height and width of 128 pixels with three channels. The output layer is made by a Conv2D convolution layer, with an activation ‘sigmoid’. The model was compiled with the adaptive moment estimation (Adam) compiler. The loss function used was binary cross-entropy, and the metric that was examined was accuracy.
The model is trained on the data with a validation split of 0.1, batch size of 16, epochs of 25, and callback. The callback was used for early stopping. The ‘val_loss’ was used to monitor.
The model was run to calculate accuracy. The accuracy function in TensorFlow calculates the frequency at which the model’s predictions match the actual labels. It does this by creating two local variables called “total” and “count”. “Total” keeps track of the total number of correct predictions, while “count” keeps track of the total number of predictions made by the model. The accuracy function then divides “total” by “count” to get the overall frequency of correct predictions, which is returned as the accuracy of the model. This is an idempotent operation, which means that it does not change the state of the model or its inputs. As summarized in Table 1, the U-net got an accuracy of 82.41% for an image size of 128×128 and 86.22% for an image size of 256×256. The Attention U-net got an accuracy of 82.14% for an image size of 128×128 and 86.35% for an image size of 256×256. The Attention residual U-net got an accuracy of 89.35% for an image size of 128×128 and 98.35% for an image size of 256×256. The difference in the performance of U-net and Attention U-net is negligible while the difference between Attention residual U-net and Attention U-net is large.
| Table 1. Accuracy values of the various methods with image sizes 128×128 and 256×256. | ||
|
Accuracy (using 128x 128 image size) |
Accuracy (using 256x 256 image size) |
|
| U-Net | 82.41 | 86.22 |
| Attention U-net | 82.43 | 86.35 |
| Att-Res Net | 89.35 | 98.35 |
When we are using a different image size, U-net and Attention U-net perform better for the larger image size. The Attention residual U-net gives the best value when we are using the 256×256 image size. Overall, the Attention residual U-net is performing better in both image sizes.
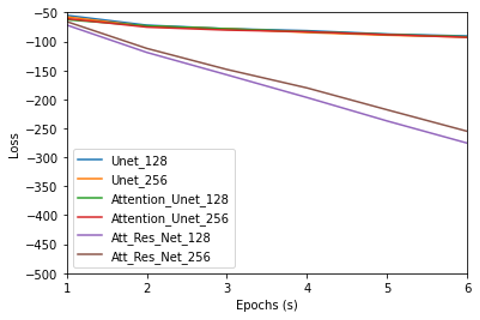
We are using binary cross-entropy loss for the selected methods. During the training, after every epoch, the loss value should decrease because the learning is increasing. As you can notice in Figure 1, Y-axis is the loss value and the x-axis is the number of epochs. In all the methods when we use the image size of 128×128, the loss of the attention residual U-net is quite low when compared to the attention U-net. When we use an image size of 256×256, the same pattern can be seen as found in the 128×128 image. The Attention residual U-net performs better when the image size is 256×256 compared to 128×128.
Figure 1. Trend in the Binary Cross Entropy Loss over a number of epochs
Although all the methods selected have an encoder-decoder setup, the reason why the Attention Residual U-net is performing better than the other methods for the 256×256 image size is that the higher resolution image has more location information. We are interested in the location information for the cancer cells location as we are looking for the exact position of the cancer cells.
CONCLUSION.
The goal of the study is to determine how adding the residual connection and attention and attention module to the U-net affects breast cancer segmentation. In this study, we evaluated U-net, attention U-net and residual attention U-net and compared the performance using accuracy. Overall, this study proved that residual attention U-net trained on the images can accurately segment the cancerous cells. This should help pathologists with breast cancer detection without the time-consuming and laborious job of manual histopathological analysis that in turn leads to improved patient care.
ACKNOWLEDGMENTS.
Thank you for the guidance of Hina Ajmal from University of Oxford in the development of this research paper.
REFERENCES.
1 Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2020. CA. Cancer J. Clin. 70, 7–30 (2020).
2 He, L., Long, L.R., Antani, S. and Thoma, G.R., Local and global Gaussian mixture models for hematoxylin and eosin stained histology image segmentation. 10th International Conference on Hybrid Intelligent Systems. 223-228 (2010).
3 Bera, K., Schalper, K.A., Rimm, D.L., Velcheti, V. and Madabhushi, A. Artificial intelligence in digital pathology—new tools for diagnosis and precision oncology. Nature reviews Clinical oncology, 16(11), 703-715 (2019).
4 Dimitriou, N., Arandjelović, O. and Caie, P.D., Deep learning for whole slide image analysis: an overview. Frontiers in medicine, 6, 264 (2019).
5 Wu, X., Sahoo, D. and Hoi, S.C., Recent advances in deep learning for object detection. Neurocomputing, 396, 39-64 (2020).
6 Gu, J., Wang, Z., Kuen, J., Ma, L., Shahroudy, A., Shuai, B., Liu, T., Wang, X., Wang, G., Cai, J. and Chen, T., Recent advances in convolutional neural networks. Pattern recognition, 77, 354-377 (2018).
7 Long, J., Shelhamer, E. and Darrell, T., Fully convolutional networks for semantic segmentation. 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 3431-3440 (2015)
8 Alom, M.Z., Yakopcic, C., Hasan, M., Taha, T.M. and Asari, V.K., Recurrent residual U-Net for medical image segmentation. Journal of Medical Imaging, 6(1), 014006-014006 (2019).
9 Dong, H., Yang, G., Liu, F., Mo, Y. and Guo, Y., Automatic brain tumor detection and segmentation using U-Net based fully convolutional networks. Medical Image Understanding and Analysis: 21st Annual Conference, MIUA 2017, Edinburgh, UK, July 11–13, 2017, Proceedings 21. 506-517. Springer International Publishing (2017)
10 Zhang, Z., Liu, Q. and Wang, Y., Road extraction by deep residual u-net. IEEE Geoscience and Remote Sensing Letters, 15(5), .749-753 (2018).
11 Zhang, Z., Wu, C., Coleman, S. and Kerr, D., DENSE-INception U-net for medical image segmentation. Computer methods and programs in biomedicine, 192, .105395 (2020).
12 Oktay, Ozan, Jo Schlemper, Loic Le Folgoc, Matthew Lee, Mattias Heinrich, Kazunari Misawa, Kensaku Mori,. Attention u-net: Learning where to look for the pancreas. Proceedings of the Medical Imaging with Deep Learning (2018)
13 He, K., Gkioxari, G., Dollár, P. and Girshick, R., Mask r-cnn. 2017 IEEE International Conference on Computer Vision (ICCV). 2961-2969 (2017).
14 Ren, S., He, K., Girshick, R. and Sun, J., Faster r-cnn: Towards real-time object detection with region proposal networks. Advances in neural information processing systems, 28, 91-99 (2015).
15 Khened, M., Kollerathu, V.A. and Krishnamurthi, G., Fully convolutional multi-scale residual DenseNets for cardiac segmentation and automated cardiac diagnosis using ensemble of classifiers. Medical image analysis, 51, 21-45 (2019).
16 Lee, C.Y., Xie, S., Gallagher, P., Zhang, Z. and Tu, Z., February. Deeply-supervised nets. Proceedings of Machine Learning Research. 38, 562-570 (2015)
17 Wolz, R., Chu, C., Misawa, K., Fujiwara, M., Mori, K. and Rueckert, D., Automated abdominal multi-organ segmentation with subject-specific atlas generation. IEEE transactions on medical imaging, 32(9), 1723-1730 (2013).
18 Long, J., Shelhamer, E. and Darrell, T., Fully convolutional networks for semantic segmentation. 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 3431-3440 (2015).
19 Ronneberger, O., Fischer, P. and Brox, T., U-net: Convolutional networks for biomedical image segmentation. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015. 18, 234-241 (2015)
20 Bai, W., Sinclair, M., Tarroni, G., Oktay, O., Rajchl, M., Vaillant, G., Lee, A.M., Aung, N., Lukaschuk, E., Sanghvi, M.M. and Zemrak, F., Automated cardiovascular magnetic resonance image analysis with fully convolutional networks. Journal of Cardiovascular Magnetic Resonance, 20(1), 1-12. (2018)
21 Roth, H.R., Lu, L., Lay, N., Harrison, A.P., Farag, A., Sohn, A. and Summers, R.M., Spatial aggregation of holistically-nested convolutional neural networks for automated pancreas localization and segmentation. Medical image analysis, 45, 94-107 (2018).
22 Schlemper, J., Oktay, O., Schaap, M., Heinrich, M., Kainz, B., Glocker, B. and Rueckert, D., Attention gated networks: Learning to leverage salient regions in medical images. Medical image analysis, 53, 197-207 (2019).
23 Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A.N., Kaiser, Ł. and Polosukhin, I., Attention is all you need. Advances in neural information processing systems, 30. (2017).
24 He, K., Zhang, X., Ren, S. and Sun, J., Deep residual learning for image recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 770-778 (2016).
25 Li, B. and Lima, D., Facial expression recognition via ResNet-50. International Journal of Cognitive Computing in Engineering, 2, 57-64 (2021).
Posted by John Lee on Tuesday, May 30, 2023 in May 2023.
Tags: breast cancer, Deep learning, Histopathological images