Investigating the Correlation Between Phytoncide and Gut Bacteria to Maintain Strong Immunity and Brain Health of Astronauts
ABSTRACT
The aim of this research is to establish a correlation between the inhalation of phytoncide, a natural bactericide emitted by certain pine trees, and human immune and brain health through its effects on the gut microbiome and other organ systems to assess phytoncide’s ability to enhance the air on spacecraft. Through population measurement of three bacteria species after treatment with phytoncide, it can be concluded that phytoncide has the capacity to reduce the populations of opportunistic E. coli and harmful C. bolteae. Furthermore, phytoncide inhalation does not appear to affect populations of human immune cells while also changing the immune-related gene expression. Although the results are only part of a small data set, it can be concluded that a correlation exists between inhalation of phytoncide and human immune health through the gut microbiome. Phytoncide has the potential to enhance air on spacecraft.
INTRODUCTION.
Many scientists are studying the possibility of migration to other planets. Thus, solutions to problems in deep-space exploration are being studied. Air in closed environments is purified to allow for easy breathing. However, bioaerosols, biological components including bacteria, viruses, and their metabolites present in the Earth’s air, have been largely ignored by researchers in regards to spaceflight. As such, little research on the impact of bioaerosol absence on human health, such as decline of immune function, in closed, sterile environments has been done [1]. Researchers have observed phytoncide’s ability to increase the number of natural killer cells present in the human body [2]. Research on the gut microbiome has been prevalent in recent years for its relationship to human health. Influencing mental [3] and physical [4] health, the microbiome has become important in improving the overall health [5][6]. These elements were investigated to see how phytoncide could impact the gut microbiome and how resulting metabolites from microbiota being exposed to phytoncide could augment peoples’ health in artificial habitats in space.
MATERIALS AND METHODS.
-
1. Preparation of Media for Gut Bacteria Culture. De Man, Rogosa and Sharpe broth (MRS), nutrient broth (NB), and Clostridium broth (CB) were made using a broth powder dissolved and heated in distilled water. The agar plates of the broth variants were made by adding 1.5 % of the volume of water in agar powder in terms of weight.
2. Cultivation of Gut Bacteria and Human Somatic Cells. Lactobacillus reuteri, Clostridium bolteae, and Escherichia coli bacteria samples were obtained from the Korean Collection for Type Cultures. L. reuteri were cultured in MRS broth medium, C. bolteae in CB medium, and E. coli in NB medium for 48 hours.
SK-N-MC cells, a human brain cell line, were cultured in DMEM (+10 % FBS + 1x antibiotics). A549 cells, a human lung cell line, were also cultured in the same solution. Daudi cells, a human immune cell line, were cultured in DMEM (+20 % FBS + 1x antibiotics). All were placed in an incubator at 37 ℃ and 5 % CO2.
3. Treatment of gut bacteria and A549 cells with Phytoncide.
3.1. Treatment of Gut Bacteria. Various dilutions of phytoncide were added to 15 mL conical tubes containing MRS, NB, or CB broth. Each bacteria strain (L. reuteri, C. bolteae, and E. coli) was cultured in separate broth samples, for a total of 9 experimental conditions with different phytoncide dilutions for each broth type. After two days in an incubator at 37 ℃, all 27 samples were placed into a microplate to measure the samples’ optical density for the changes in population due to the addition of phytoncide.
3.2. Treatment of A549 Cells. 100 µL of two concentrations of diluted phytoncide, 25% and 6.25% phytoncide by volume, were injected into T-25 flasks, labeled for the concentrations of phytoncide injected into them, with one T-25 flask used to cultivate the negative control A549 cells that did not receive any phytoncide treatment. After three days, the media and the metabolites in them were extracted from each of the flasks using a syringe filter. Given that metabolites that could impact human health have to be expunged from the cell’s inner environment into the outer medium, extraction of media ensured that any significant metabolites affecting human health are also extracted.
4. Extraction of Metabolites from A549 Cells and Gut Bacteria. The three bacteria strains, L. reuteri, C. bolteae, and E. coli, were mixed in a 15 ml tube at a 1:1:1 ratio along with either 1 µl, 5 µl, or 10 µl of A549 metabolite, resulting in a set of three 15 ml tubes for every phytoncide concentration, including the negative control, used at the A549 metabolite creation stage, with one tube of bacteria mixed in without A549 metabolites of any kind. After mixing, the mixtures were left to incubate at 37 ˚C for 4 days, after which the mixtures were transferred to 1.5 mL tubes. After the selection streaking (see next section), all tubes were spun in a centrifuge at 13,500 RPM for 15 minutes to form the supernatant layer with the pellet of bacteria at the bottom. The pellets were disposed while the supernatant liquids from each sample were kept, as it now contained the metabolites of the bacteria.
5. Gut Bacteria Selection Streaking. Petri dishes were prepared to selectively streak for specific bacteria and compare the sizes and numbers of colonies. The samples of mixed gut microbiota, before going to the centrifuge, were streaked onto MRS agar, NA, and Clostridium agar plates. The bacteria strains on each plate were identified using the media they were on, and the size and number of colonies on the plates were observed.
6. Reverse-Transcriptase Polymerase Chain Reaction (RT-PCR).
6.1. Isolation of total RNA. First, the media was taken out of the wells. For SK-N-MC, the cells were taken off the well’s bottom with a 0.25 % trypsin/EDTA solution, and the contents were diluted with DPBS. For both cell strains, the cells in their media were moved to a 1.5 ml tube and spun at 1,500 RPM for 5 minutes. The media was then removed, leaving only the pellet. RB buffer was then added, and the contents were homogenized in a vortex mixer. 80 % ethanol was then added and immediately mixed with a pipette. The contents were then transferred to binding columns and collection tubes. The tubes were spun at 12,000 RPM for 1 minute. The contents of the collection tubes were emptied, and RWA2 buffer was added into the binding columns. The tubes were spun at 12,000 RPM for 1 minute. The contents of the collection tubes were emptied, and RWA2 buffer was added into the binding columns. This time, the tubes were spun at 12,000 RPM for 5 minutes. After emptying the collection tubes, the tubes were spun with nothing in the binding columns at 12,000 RPM for 2 minutes. The contents of the collection tubes were emptied, and the collection tubes were replaced with new 1.5 ml tubes. ER buffer solution was then added and kept in the binding columns for 1 minute. The tubes were spun in a centrifuge at 10,000 RPM for 1 minute to allow the cells to elute.
6.2. cDNA synthesis. The cells’ extracted RNA was put through reverse transcription to obtain cDNA. The RNA was transferred into ten PCR tubes. From there, a mixture with Oligo(dT) primer to RTase to 5x buffer to water ratio of 0.5:0.2:2:2.3 was made, and 5µl of the solution was added to each tube. The PCR tubes were then placed into a thermal cycler to perform the RT process at 42 ˚C for 1 hour. The contents of the PCR tubes were then transferred to 1.5 ml tubes and distilled water was added to dilute the contents.
6.3. PCR. For PCR, the process to extrapolate gene expression in cells, mixtures of forwards and backwards gene primers were made with distilled water. A 30:20:20:20:2 ratio of Daudi cDNA to primer solution to dNTP to 10x buffer solution to TAQ polymerase was mixed with enough water for the water to make up 54 % of the mixture’s entire volume. This was run through the thermal cycler to amplify the quantity of a section of the GAPDH. The cycle was the following: 94 ˚C for 30 seconds, then a cycle of 94 ˚C for 15 seconds, 60 ˚C for 15 seconds, and 72 ˚C for 15 seconds, moving on to 72 ˚C for 10 seconds, 10 ˚C for storage indefinitely.
The agarose gel was made by heating agarose powder and 1xTAE buffer in a microwave until thoroughly dissolved. This was poured into the gel electrophoresis system trays. The tubes were then removed, and its contents were placed into a gel electrophoresis system. The systems were run from between 25-30 minutes, depending on the length of the protein. The agarose gel was removed from the systems after time was up and placed under a UV illuminator in a Gel Doc, and the results were documented.
RESULTS.
1. Effect of Phytoncide on Gut Bacteria Growth. The results of the optical density measurements of the beneficial intestinal bacteria L. reuteri [5] and the harmful bacteria C. bolteae [6] show insignificant variation in optical density. However, the opportunistic bacteria E. coli [7] optical density measurements show a negative relationship between the amount of phytoncide in the medium and the measured optical density. In other words, with the addition of more phytoncide, the optical density of the E. coli sample decreases exponentially, indicating a lower density of bacteria in the medium (Fig. 1). Thus, the addition of phytoncide appears to be able to inhibit the growth of E. coli while maintaining the population of L. reuteri and C. bolteae.
Figure 1. Growth of L. reuteri, C. bolteae and E. coli after treatment with Phytoncide
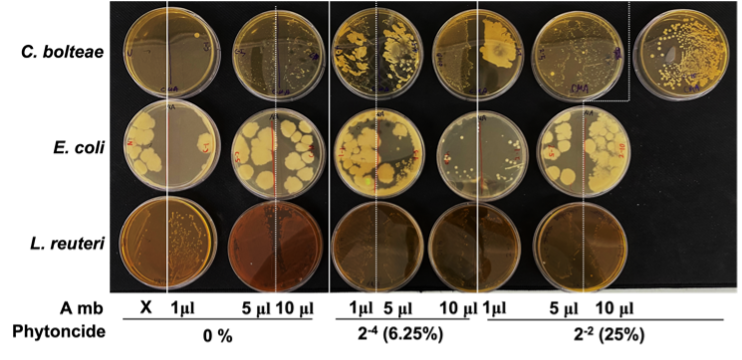
For each respective concentration of phytoncide, the C. bolteae colonies became more numerous as more A549 metabolite was added into the media of the bacteria. However, the general trend across all groups shows that the bacterial colonies seemed to decrease in number as phytoncide concentration went up within the A549 metabolites added into the media. In E. coli colonies, colony size generally increased with more addition of A549 metabolites as well as with greater phytoncide concentrations within each A549 metabolite variant. Colony numbers for E. coli did not change in a significant way for the most part. L. reutericolonies appeared to increase in number but remained relatively unchanged in their size (Fig. 2). In all, with greater presence of phytoncide within A549 metabolites, C. bolteae colony numbers fell, E. coli colony numbers stayed the same, and L. reuteri colony numbers increased.
Figure 2. Formation of gut bacteria colonies after treatment with phytoncide-treated A549 cell metabolite. (A, A549 cells; mb, metabolite)
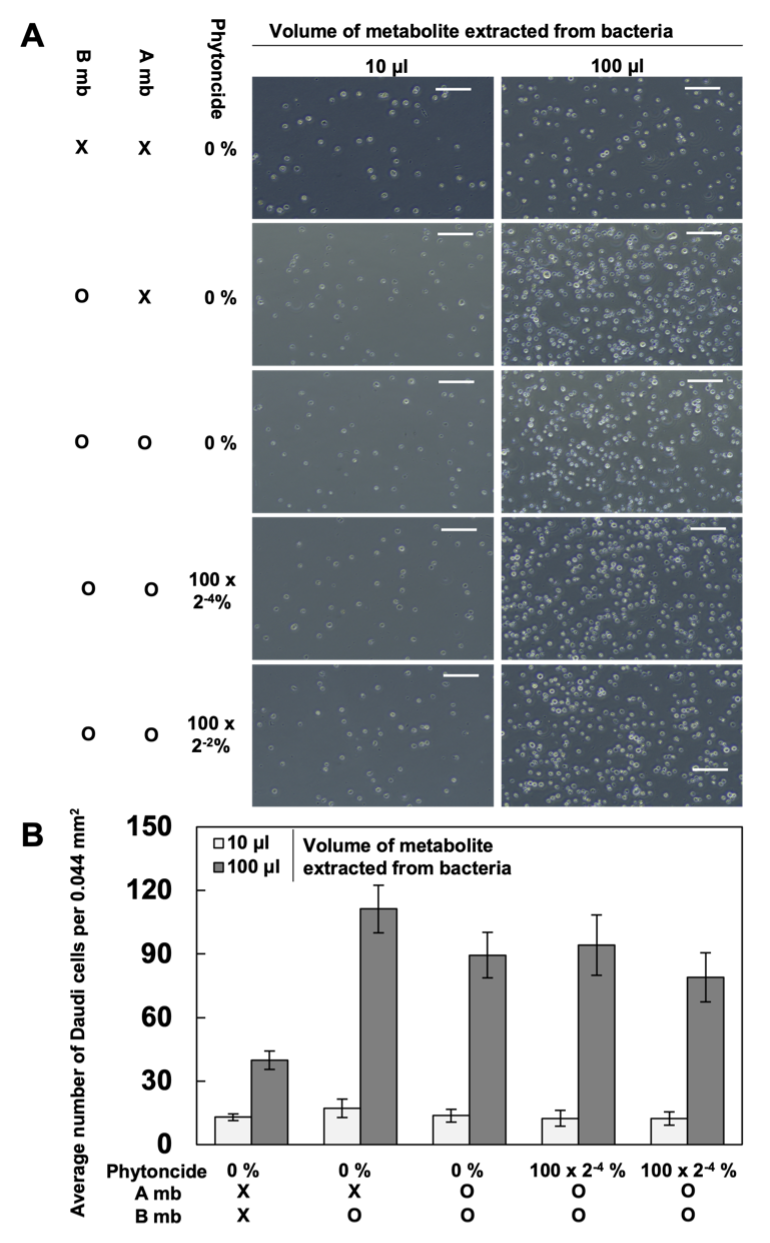
2. Effects of Gut Bacteria Metabolites Treated with Metabolites of Phytoncide-treated A549 Cells on SK-N-MC and Daudi Cells. Based on the individual cell counting of the samples created after treating the Daudi cells with the intestinal flora metabolites (Fig. 3), the Daudi cells have no significant changes in cell density with or without phytoncide. It is evident, however, that Daudi cell density increases by a considerable amount, visually speaking, when bacteria metabolites are added in greater quantity in all the sets.
Figure 3. Daudi cells after treatment with gut bacteria metabolites (X, Untreated; B, Treated with gut bacteria metabolite; P, Phytoncide; O, Treated cells; A, A549 cells; mb, A549 metabolite. Bars mean size bar (=100μm). A. The captured images; B. The average number of Daudi cells)
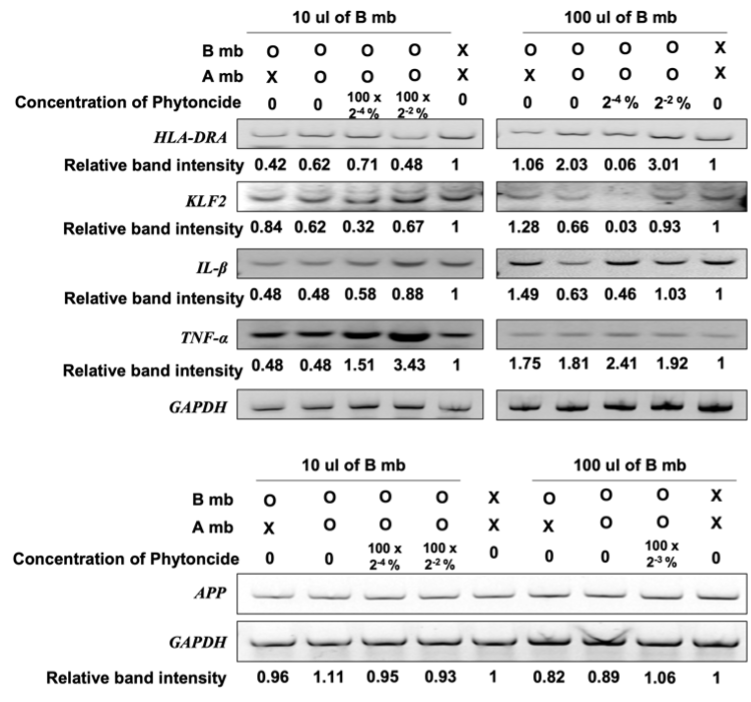
3. Expression of Daudi and SK-N-MC Cell Genes After Exposure to Metabolites. The metabolites of the intestinal flora, after treatment with the metabolites extracted from phytoncide-treated A549 cells, were used to treat Daudi cells. The changes in the expression of HLA-DRA, a gene that aids in antigen presenting in immune cells [8], KLF2, a gene that helps differentiate and maintain T-lymphocyte cells [9], IL-β, whose overexpression contributes to the development of cancer through pro-cancer inflammation [10], and TNF-α, a gene that serves as an important signaling molecule for autoimmunity-associated pro-inflammatory macrophages [11] in the treated Daudi cells were investigated.
Previous studies have shown that changes in the balance of gut bacteria are correlated to incidence of Alzheimer’s [12] and that phytoncides can help maintain brain health. To this end, the metabolites of gut bacteria treated with the metabolites of lung cells exposed to phytoncide were used to treat SK-N-MC cells, a human neuronal cell line. Then, the expression changes of the APP gene, which is an Alzheimer’s-inducing protein, was recorded.
In a 10 µL gut bacteria metabolite-present environment, the deviation of APP gene relative band intensity does not exceed ±0.11 in any of the environmental variations in which the expression of APP in SK-N-MC cells was measured in. In the 100 µL gut bacteria metabolite-present environment, the deviation of APPgene relative band intensity also does not exceed 0.18 in all measured cases in which APP expression was measured for in. The small difference in sample relative band intensity deviation between the 10 µL gut bacteria metabolite-present environment samples and 100 µL gut bacteria metabolite-present environment samples is despite a ten-fold increase in the amount of bacteria metabolite present in the environment, suggesting insignificance in the differences of relative band intensity values between all the samples and their comparative standards.
Figure 4. Relative gene expression of genes in Daudi cells and SK-N-MC cells involved in regulation of immunity and brain function respectively (X, Untreated cells; B, Cells treated with metabolite gut bacterium that have not been treated with metabolite from A549; P, Phytoncide; O, Treated cells; A, A549 cells; mb, metabolite from A549 cells. The band intensity of each gene was normalized by the intensity of GAPDH, and the relative intensity was compared with the untreated cells.)
DISCUSSION.
The rushed period in which the experimentation was performed resulted in a small data set from which conclusions were drawn. As such, the patterns observed in the cells may be more exaggerated or underrepresented than in reality. A number of confounding variables were overlooked due to a lack of time, where scale of experimentation led to a lack of feasible repeatability for experimentation within the allotted time given for experimentation. This led to a small data set that unfortunately led to a lack of clear patterns to follow and thus a deficiency of clear conclusions to take away from said experimentation. General trends were hard to read, and guaranteeing verity from a data set that could have been skewed by unfortunate experimentation conditions and random chance is difficult. However, the observed changes were still likely linked to the addition of metabolites and phytoncide, so the conclusions in this paper should be investigated further to test for the extent of effect. For future experimentation, a greater focus on one aspect of this niche is advised, as the complexity of certain parts of experimentation must not be underestimated.
CONCLUSION.
In short, based on the observed decreases in E. coli and C. bolteae populations when exposed to phytoncide in the form of decreased optical density with suspended E. coli and lower colony numbers with the C. bolteae on the petri dish, it can tentatively be concluded that the inhalation of phytoncide into the lungs and subsequent absorption into the body’s microbiome can generally improve the health of said microbiome through reduction of opportunistic bacteria and harmful bacteria populations. We are currently unable to make a conclusion on the effect of affected bacteria metabolites on the health of both immune cells or brain cells due to inconclusive results from the cell counting of Daudi cells, yielding no differences in cell population despite the addition of phytoncide, and the lack of significant deviation in terms of gene expression for SK-N-MC cells.
ACKNOWLEDGMENTS.
I would like to thank my research supervisor, Myoung-Hoon Lee, for teaching me the ropes to operating in the lab environment.
REFERENCES.
[1] D. S. Levine, J. E. Greenleaf, Immunosuppression during spaceflight deconditioning. Aviat. Space Environ. Med. 69, 172-177 (1998).
[2] Q. Li, M. Kobayashi, Y. Wakayama, H. Inagaki, M. Katsumata, Y. Hirata, K. Hirata, T. Shimizu, T. Kawada, B. J. Park, T. Ohira, T. Kagawa, Y. Miyazaki, Effect of phytoncide from trees on human natural killer cell function. Int. J. Immunopathol. Pharmacol. 22, 951-959 (2009).
[3] L. G. Canipe, M. Sioda, C. L. Cheatham, Diversity of the gut-microbiome related to cognitive behavioral outcomes in healthy older adults. Arch. Gerontol. Geriatr. 96, (2021).
[4] C. M. F. Snethlage, M. Nieuwdorp, D. H. V. Raalte, E. Rampanelli, B. C. Verchere, N. M. J. Hanssen, Auto-immunity and the gut microbiome in type 1 diabetes: Lessons from rodent and human studies. Best Pract. Res. Clin. Endocrinol. Metab. 35, (2021).
[5] A. A. Voorhies, C. Mark Ott, S. Mehta, D. L. Pierson, B. E. Crucian, A. Feiveson, C. M. Oubre, M. Torralba, K. Moncera, Y. Zhang, E. Zurek, H. A. Lorenzi, Study of the impact of long-duration space missions at the International Space Station on the astronaut microbiome. Sci. Rep. 9, (2019).
[6] A. S. Wilson, K. R. Koller, M. C. Ramaboli, L. T. Nesengani, S. Ocvirk, C. Chen, C. A. Flanagan, F. R. Sapp, Z. T. Merritt, F. Bhatti, T. K. Thomas, S. D. J. O’Keefe, Diet and the Human Gut Microbiome: An International Review. Dig. Dis. Sci. 65, (2020).
[5] J. Walter, Ecological role of lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Appl. Environ. Microbiol. 74, 4985–4996 (2008).
[6] Y. Song, C. Liu, D. R. Molitoris, T. J. Tomzynski, P. A. Lawson, M. D. Collins, S. M. Finegold, Clostridium bolteae sp. nov., Isolated from Human Sources. Syst. Appl. Microbiol. 26, 84-89 (2003).
[7] J. N. V. Martinson, S. T. Walk, Escherichia coli Residency in the Gut of Healthy Human Adults. EcoSal Plus 9, (2020).
[8] M. A. Blanar, E. C. Boettger, R. A. Flavell, Transcriptional activation of HLA-DR alpha by interferon gamma requires a trans-acting protein. Proc. Natl. Acad. Sci. U.S.A.85, 4672-4676 (1988).
[9] C. M. Carlson, B. T. Endrizzi, J. Wu, X. Ding, M. A. Weinreich, E. R. Walsh, M. A. Wani, J. B. Lingrel, K. A. Hogquist, S. C. Jameson, Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature 442, 299–302 (2006).
[10] N. G. Díaz-Maroto, G. Garcia-Vicién, G. Polcaro, M. Banuls, N. Albert, A. Villanueva, D. G. Mollevi, The Blockade of Tumoral IL1β-Mediated Signaling in Normal Colonic Fibroblasts Sensitizes Tumor Cells to Chemotherapy and Prevents Inflammatory CAF Activation. Int. J. Mol. Sci. 22, 4960 (2021).
[11] R. R. El-Tahan, A. M. Ghoneim, N. El-Mashad, TNF-α gene polymorphisms and expression. Springerplus 5, 1-7 (2016).
[12] C. Jiang, G. Li, P. Huang, Z. Liu, B. Zhao, The Gut Microbiota and Alzheimer’s Disease. J. Alzheimers. Dis. 58, 1-15 (2017).
Posted by John Lee on Tuesday, May 30, 2023 in May 2023.
Tags: Immunity, Microbiome, Phytoncide, Spacecraft