Research
Phylogenomics
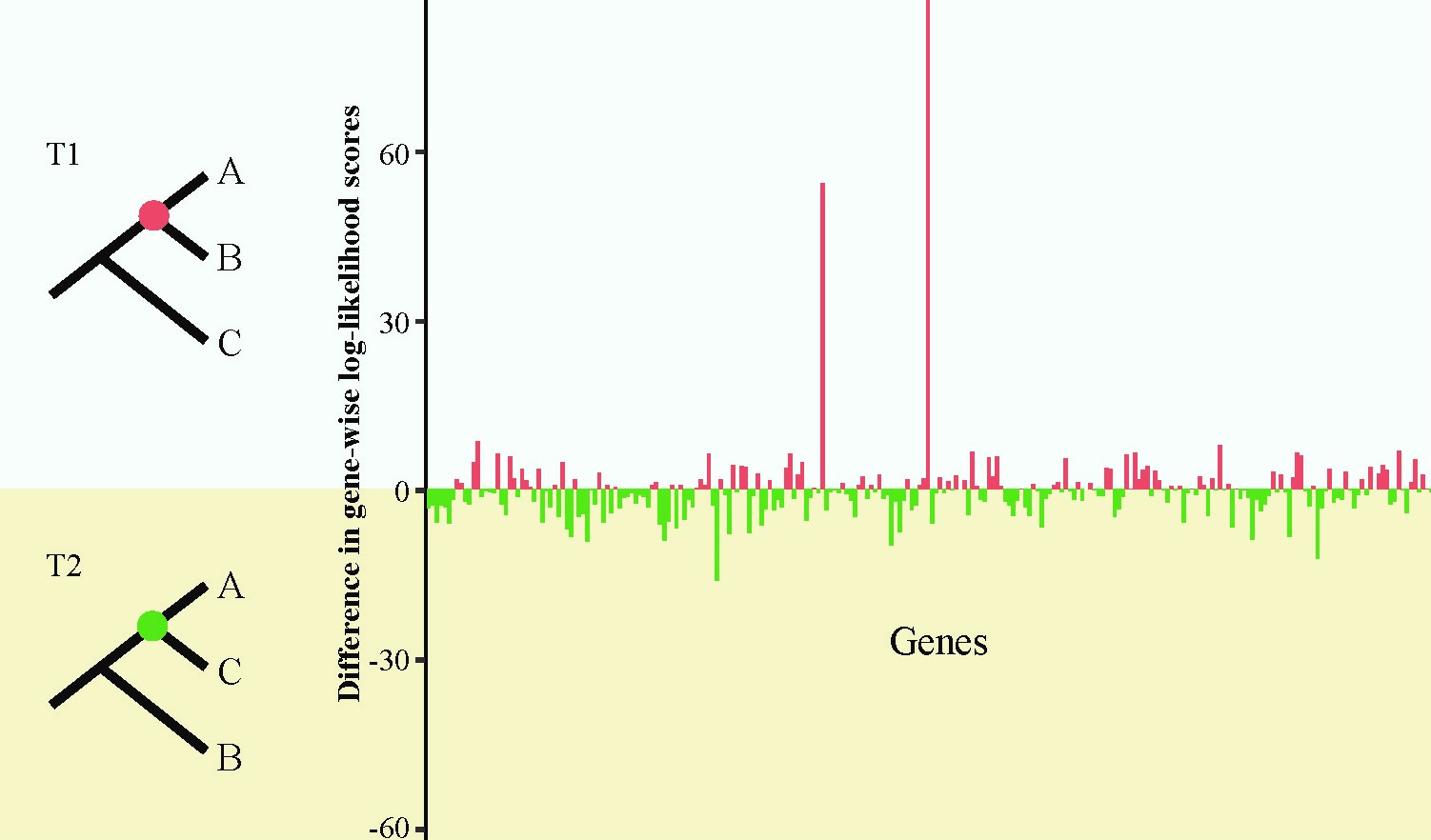
Understanding the factors that contribute to phylogenetic inaccuracy is critical for the generation of robust phylogenies, for inferring the evolution of genes, pathways, and traits, and for the testing of evolutionary hypotheses. We use diverse sets of phylogenomic data from animals, plants, and especially fungi to investigate the parameters affecting phylogenetic accuracy, develop assays for the detection of conflicts in molecular phylogenies, and examine the effect of homoplasy and selection on phylogenetic estimation.
Representative publications:
Zhou et al. (2018) Mol. Biol. Evol.
Shen et al. (2017) Nature Ecol. Evol.
Salichos & Rokas (2013) Nature
Evolution of Fungal Primary and Secondary Metabolism
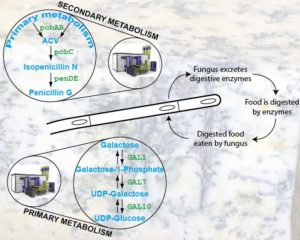
A defining characteristic of Fungi is that, in contrast to animals, they are typically embedded in their food and digest it externally in the presence of competitors (think of the blue lines of mold in blue cheese). Thus, different fungi specialize in “eating” different foods, hence their diverse primary metabolism. In collaboration with the Hittinger lab at the University of Wisconsin-Madison and with the late pioneer of budding yeast taxonomy Clete Kurtzman, we are sequencing and analyzing the genomes of all known yeast species from the subphylum Saccharomycotina to study the evolution of their diverse metabolic and ecological functions. For more information, please check the Y1000+ Project website.
Representative publications:
Steenwyk et al. (2019) PLoS Biol.
Kominek et al. (2019) Cell
Shen et al. (2018) Cell
Because digestion happens externally, fungi have also evolved potent food defense mechanisms (hence their diverse secondary metabolism). The cholesterol-reducing drug lovastatin, the antibiotic penicillin, and the potent carcinogen aflatoxin are just a tiny sample of the pharmacopoeia stored in and manufactured by fungal DNA. Our lab studies elucidate how this diversity is generated and maintained as well as how it contributes to fungal pathogenicity. For these studies, we largely study Aspergillus molds and we are collaborating with Gustavo Goldman’s lab at the University of Sao Paolo and with Nick Oberlies’ lab at the University of North Carolina at Greensboro.
Representative publications:
Mead et al. (2019) mSphere
Rokas et al. (2018) Nature Rev. Microbiol.
Lind et al. (2017) PLoS Biol.
Evolution of Human Pregnancy
Evolutionists have long been fascinated with how traits as complex and yet so closely tied to evolutionary fitness as pregnancy-related ones could have evolved. Recent discoveries stemming from comparisons with our closest living relatives, the chimpanzees, as well as other extinct hominins are revealing unmistakable signatures of the dramatically fast evolution of many pregnancy-related traits in our genomes, skeletons, brains, behaviors, and cultures. Perhaps less obvious, but no less important, is the story of how modern human pregnancy, much like the modern human body, evolved out of deeper ancestral vertebrate forms. We are working on a wide variety of research questions surrounding human pregnancy aiming to generate a synthetic portrait of its evolution. In parallel, we are aiming to understand the forces that have sculpted the recent evolution of loci associated with one of the major complications of human pregnancy, preterm birth.
Representative publications:
Lamm et al. (2018) eLife
Eidem et al. (2018) BMC Medical Genomics
Abbot & Rokas (2017) Curr. Biol.